Gabriel Brammer
(This page is auto-generated from the Jupyter notebook photometric-catalog-demo.ipynb.)
Show how to interact with the DJA/grizli photometric catalogs.
(little explanatory text for the quick demo)
%matplotlib inline
import os
import yaml
import numpy as np
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings('ignore')
import astropy.io.fits as pyfits
import grizli
import grizli.catalog
from grizli import utils
import eazy
print(f'grizli version: {grizli.__version__}')
print(f'eazy-py version: {eazy.__version__}')
grizli version: 1.10.dev2+g661e5ea
eazy-py version: 0.6.5
Set the field
Currently available:
gds= GOODS-Southgdn= GOODS-Northceers-full= CEERS EGSabell2744clu= Abell 2744 GLASS + UNCOVER + DD-2756macs0647= MACS 0647 cluster (Coe et al., GO-1433)rxj2129= RXJ 2129 cluster (Kelly et al., DD-2767)sunrise= “Sunrise Arc” (WHL0137, Coe et al., GO-2282)smacs0723= SMACS 0723 cluster (Pontoppidan et al., DD-2736)- …
field = 'gds-grizli-v7.0'
url_path = 'https://s3.amazonaws.com/grizli-v2/JwstMosaics/v7'
Raw photometry
NB: All photometry given in fnu flux densities with units of microJansky (AB zeropoint = 23.9).
phot = utils.read_catalog(f'{url_path}/{field}_phot.fits')
Metadata
# General data of the source detection
for i, k in enumerate(phot.meta):
print(f'{k:>36} = {phot.meta[k]}')
if i > 70:
print('...')
break
VERSION = 1.2.1
MINAREA = 9
CLEAN = True
DEBCONT = 0.001
DEBTHRSH = 32
FILTER_TYPE = conv
THRESHOLD = 1.5
KRONFACT = 2.5
KRON0 = 2.4
KRON1 = 3.8
MINKRON = 8.750000000000059
TOTCFILT = F140W
TOTCWAVE = 13922.907
ZP = 28.9
PLAM = 13922.907
FNU = 1e-08
FLAM = 1.4737148e-20
UJY2DN = 99.99395614709495
DRZ_FILE = gds-grizli-v7.0-ir_drc_sci.fits.gz
WHT_FILE = gds-grizli-v7.0-ir_drc_wht.fits.gz
GET_BACK = True
BACK_BW = 50
BACK_BH = 50
BACK_FW = 3
BACK_FH = 3
BACK_PIXEL_SCALE = 0.04
ERR_SCALE = 0.8123676180839539
RESCALEW = True
APERMASK = True
GAIN = 2000.0
APER_0 = 9.00000000000006
ASEC_0 = 0.36
APER_1 = 12.50002500000008
ASEC_1 = 0.5000009999999999
APER_2 = 17.50000500000012
ASEC_2 = 0.7000002000000001
APER_3 = 25.00000500000017
ASEC_3 = 1.0000002
CLEARP-F430M_VERSION = 1.2.1
CLEARP-F430M_ZP = 28.9
CLEARP-F430M_PLAM = 42816.84172704966
CLEARP-F430M_FNU = 1e-08
CLEARP-F430M_FLAM = 1.63095481504235e-22
CLEARP-F430M_uJy2dn = 99.99395614709495
CLEARP-F430M_DRZ_FILE = gds-grizli-v7.0-clearp-f430m_drc_sci
CLEARP-F430M_WHT_FILE = gds-grizli-v7.0-clearp-f430m_drc_wht
CLEARP-F430M_GET_BACK = True
CLEARP-F430M_BACK_BW = 50
CLEARP-F430M_BACK_BH = 50
CLEARP-F430M_BACK_FW = 3
CLEARP-F430M_BACK_FH = 3
CLEARP-F430M_BACK_PIXEL_SCALE = 0.04
CLEARP-F430M_ERR_SCALE = 1.0
CLEARP-F430M_RESCALEW = True
CLEARP-F430M_APERMASK = True
CLEARP-F430M_GAIN = 2000.0
CLEARP-F430M_aper_0 = 9.00000000000006
CLEARP-F430M_asec_0 = 0.36
CLEARP-F430M_aper_1 = 12.50002500000008
CLEARP-F430M_asec_1 = 0.5000009999999999
CLEARP-F430M_aper_2 = 17.50000500000012
CLEARP-F430M_asec_2 = 0.7000002000000001
CLEARP-F430M_aper_3 = 25.00000500000017
CLEARP-F430M_asec_3 = 1.0000002
CLEARP-F480M_VERSION = 1.2.1
CLEARP-F480M_ZP = 28.9
CLEARP-F480M_PLAM = 48151.90220745452
CLEARP-F480M_FNU = 1e-08
CLEARP-F480M_FLAM = 1.28956813045049e-22
CLEARP-F480M_uJy2dn = 99.99395614709495
CLEARP-F480M_DRZ_FILE = gds-grizli-v7.0-clearp-f480m_drc_sci
CLEARP-F480M_WHT_FILE = gds-grizli-v7.0-clearp-f480m_drc_wht
...
Photometric apertures
for i, k in enumerate(phot.meta):
if k.startswith('APER_'):
aper_index = k[-1]
print(f"Aperture index {aper_index}: *diameter* = {phot.meta[k]:4.1f} pixels = {phot.meta[k.replace('APER','ASEC')]:.2f} arcsec")
Aperture index 0: *diameter* = 9.0 pixels = 0.36 arcsec
Aperture index 1: *diameter* = 12.5 pixels = 0.50 arcsec
Aperture index 2: *diameter* = 17.5 pixels = 0.70 arcsec
Aperture index 3: *diameter* = 25.0 pixels = 1.00 arcsec
# Columns for a particular filter + aperture
aper_index = '1'
cols = []
for k in phot.colnames:
if k.startswith('f444w') & k.endswith(aper_index):
cols.append(k)
phot[cols].info()
<GTable length=52427>
name dtype unit class n_bad
-------------------------- ------- ---- ------------ -----
f444w-clear_flux_aper_1 float64 uJy MaskedColumn 640
f444w-clear_fluxerr_aper_1 float64 uJy MaskedColumn 640
f444w-clear_flag_aper_1 int16 Column 0
f444w-clear_bkg_aper_1 float64 uJy MaskedColumn 641
f444w-clear_mask_aper_1 float64 Column 0
Photometric bands
- NIRCam filters generally have “clear” in the filter name, which is the element in the
pupilwheel. - Filters that start with
clearpare generally the long-wavelength NIRISS filters. - Filters with names that end in
wnare the NIRISS versions, e.g.,f200wn-clearfor NIRISS andf200w-clearfor NIRCam - HST filters ending in “u” are the WFC3/UVIS versions, e.g.,
f814wu
count = 0
for k in phot.colnames:
if k.endswith('_flux_aper_1'):
count += 1
print(f"{count:>2} {k.split('_flux')[0]}")
1 clearp-f430m
2 clearp-f480m
3 f090w-clear
4 f105w
5 f110w
6 f115w-clear
7 f115wn-clear
8 f125w
9 f140w
10 f150w-clear
11 f150wn-clear
12 f160w
13 f182m-clear
14 f200w-clear
15 f200wn-clear
16 f210m-clear
17 f277w-clear
18 f335m-clear
19 f336wu
20 f350lpu
21 f356w-clear
22 f410m-clear
23 f430m-clear
24 f435w
25 f444w-clear
26 f460m-clear
27 f475w
28 f480m-clear
29 f606w
30 f606wu
31 f775w
32 f814w
33 f814wu
34 f850lp
35 f850lpu
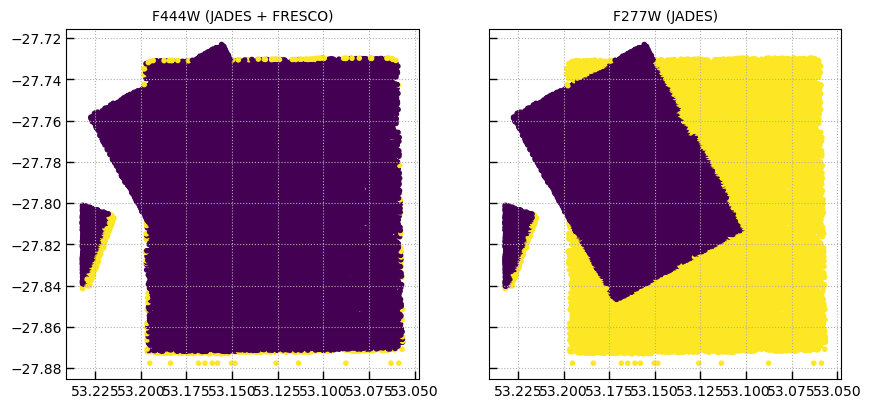
# Missing data are *masked*
fig, axes = plt.subplots(1,2,figsize=(10,5), sharex=True, sharey=True)
cosd = np.cos(np.nanmedian(phot['dec'])/180*np.pi)
axes[0].scatter(phot['ra'], phot['dec'], c=phot['f444w-clear_flux_aper_1'].mask)
axes[0].set_title('F444W (JADES + FRESCO)')
axes[1].scatter(phot['ra'], phot['dec'], c=phot['f277w-clear_flux_aper_1'].mask)
axes[1].set_title('F277W (JADES)')
axes[0].set_xlim(*axes[0].get_xlim()[::-1])
for ax in axes:
ax.set_aspect(1./cosd)
ax.grid()
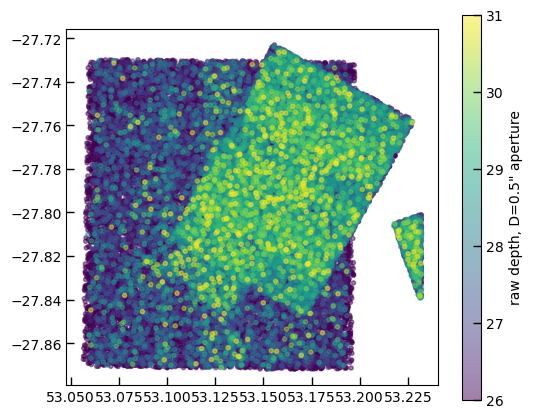
# 5-sigma depth in the D=0.5" aperture
depth = 23.9 - 2.5*np.log10(phot['f444w-clear_flux_aper_1']*5)
fig, ax = plt.subplots(1,1,figsize=(6,5))
ax.set_aspect(1./cosd)
so = np.argsort(depth)
sc = ax.scatter(phot['ra'][so], phot['dec'][so], c=depth[so], vmin=26, vmax=31, alpha=0.5)
ax.set_xlim(*axes[0].get_xlim()[::-1])
cb = plt.colorbar(sc)
cb.set_label('raw depth, D=0.5" aperture')
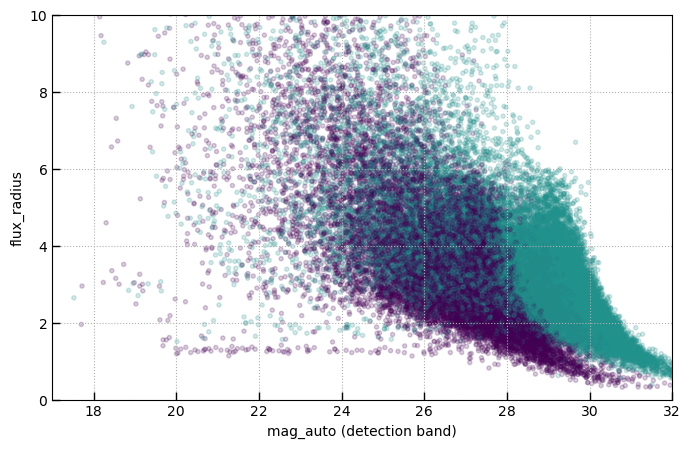
Point sources
fig, ax = plt.subplots(1,1,figsize=(8,5))
in_jades = ~phot['f277w-clear_flux_aper_1'].mask
ax.scatter(phot['mag_auto'], phot['flux_radius'], alpha=0.2, c=in_jades, vmin=0, vmax=2, cmap='viridis')
ax.set_ylim(0,10)
ax.grid()
ax.set_xlim(17, 32)
ax.set_xlabel('mag_auto (detection band)')
ax.set_ylabel('flux_radius')
Text(0, 0.5, 'flux_radius')
Catalog with aperture corrections
{filter}_corr_{aper} = {filter}_corr_{aper} * flux_auto / flux_{aper}: Aperture corrected to theautoflux in the detection band{filter}_tot_{aper} = {filter}_corr_{aper} * {filter}_tot_corr: Corrected for flux outside of the auto aperture. Not implemented for JWST, wherecorr = tot
if not os.path.exists(f'{field}-fix.photoz.tar.gz'):
! wget {url_path}/{field}-fix.photoz.tar.gz
! tar xzvf {field}-fix.photoz.tar.gz
x gds-grizli-v7.0-fix.eazypy.h5
x gds-grizli-v7.0-fix.eazypy.residuals.001.png
x gds-grizli-v7.0-fix.eazypy.residuals.002.png
x gds-grizli-v7.0-fix.eazypy.residuals.003.png
x gds-grizli-v7.0-fix.eazypy.zout.fits
x gds-grizli-v7.0-fix.eazypy.zphot.param
x gds-grizli-v7.0-fix.eazypy.zphot.translate
x gds-grizli-v7.0-fix.eazypy.zphot.zeropoint
x gds-grizli-v7.0-fix.zhist.png
x gds-grizli-v7.0-fix.zphot_zspec.png
x gds-grizli-v7.0-fix_phot_apcorr.fits
apc = utils.read_catalog(f'{field}-fix_phot_apcorr.fits')
aper_index = '1'
cols = []
for k in apc.colnames:
if k.startswith('f444w') & k.endswith(aper_index):
cols.append(k)
apc[cols].info()
<GTable length=52427>
name dtype unit class n_bad
-------------------- ------- ---- ------------ -----
f444w_flux_aper_1 float64 uJy MaskedColumn 640
f444w_fluxerr_aper_1 float64 uJy MaskedColumn 640
f444w_flag_aper_1 int16 MaskedColumn 0
f444w_bkg_aper_1 float64 uJy MaskedColumn 641
f444w_mask_aper_1 float64 Column 0
f444w_corr_1 float64 uJy Column 0
f444w_ecorr_1 float64 uJy Column 0
f444w_tot_1 float64 uJy Column 0
f444w_etot_1 float64 uJy Column 0
Compare to Skelton 3D-HST
import grizli.catalog
s14 = grizli.catalog.query_tap_catalog(ra=np.nanmedian(phot['ra']), dec=np.nanmedian(phot['dec']),
radius=10,
vizier=True,
db='"J/ApJS/214/24/3dhstall"')
len(s14)
idx, dr, dx, dy = utils.GTable(s14).match_to_catalog_sky(apc, get_2d_offset=True)
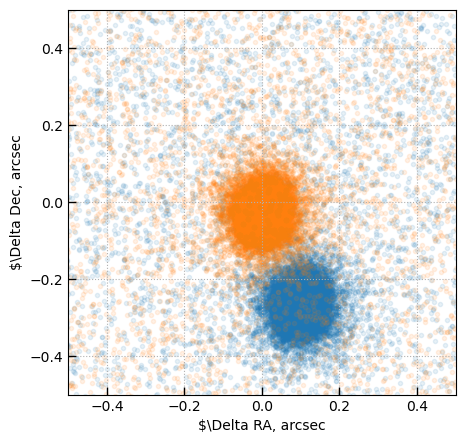
fig, ax = plt.subplots(1,1,figsize=(5,5))
ax.scatter(dx, dy, alpha=0.1)
has_match = dr.value < 0.3
ra_offset = np.nanmedian((apc['ra'] - s14['RAJ2000'][idx])[has_match])
dec_offset = np.nanmedian((apc['dec'] - s14['DEJ2000'][idx])[has_match])
s14['ra'] += ra_offset
s14['dec'] += dec_offset
idx, dr, dx, dy = utils.GTable(s14).match_to_catalog_sky(apc, get_2d_offset=True)
ax.scatter(dx, dy, alpha=0.1)
has_match = dr.value < 0.2
ax.set_xlim(-0.5, 0.5)
ax.set_ylim(*ax.get_xlim())
ax.grid()
ax.set_xlabel(r'$\Delta RA, arcsec')
ax.set_ylabel(r'$\Delta Dec, arcsec')
Query "J/ApJS/214/24/3dhstall" from VizieR TAP server
Launched query: 'SELECT TOP 1000000 * FROM "J/ApJS/214/24/3dhstall" WHERE RAJ2000 > 53.05300162385859 AND RAJ2000 < 53.24140686277677 AND DEJ2000 > -27.87878381298912 AND DEJ2000 < -27.712117146322456 '
------>http
host = tapvizier.u-strasbg.fr:80
context = /TAPVizieR/tap/sync
Content-type = application/x-www-form-urlencoded
200 200
[('date', 'Fri, 14 Jul 2023 22:54:49 GMT'), ('server', 'Apache/2.4.41 (Ubuntu) mod_jk/1.2.46 OpenSSL/1.1.1f'), ('vary', 'Accept-Encoding'), ('access-control-allow-origin', '*'), ('access-control-allow-credentials', 'true'), ('transfer-encoding', 'chunked'), ('content-type', 'application/x-votable+xml; serialization=TABLEDATA;charset=UTF-8')]
Retrieving sync. results...
Saving results to: sync_20230715005449.xml
Query finished.
Text(0, 0.5, '$\\Delta Dec, arcsec')
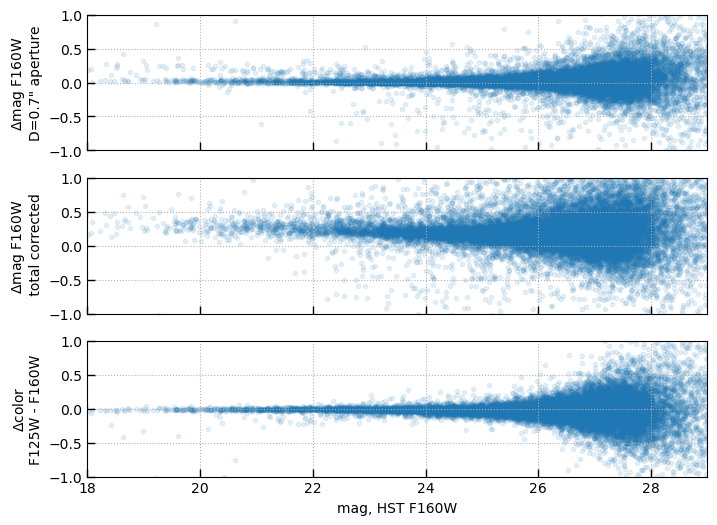
mag_s14_aper = 25 - 2.5*np.log10(s14['F160Wap'])
mag_jw_d0p7 = 23.9 - 2.5*np.log10(phot['f160w_flux_aper_2'])
mag_s14_tot = 25 - 2.5*np.log10(s14['F160W'])
mag_jw_tot = 23.9 - 2.5*np.log10(apc['f160w_tot_1'])
delta_mag_aper = mag_jw_d0p7 - mag_s14_aper[idx]
delta_mag_tot = mag_jw_tot - mag_s14_tot[idx]
# jh_jw = -2.5*np.log10(apc['f814w_tot_1']*0.847/apc['f160w_tot_1'])
# jh_s14 = -2.5*np.log10(s14['F814W']/s14['F160W'])
jh_jw = -2.5*np.log10(apc['f125w_tot_1']/apc['f160w_tot_1'])
jh_s14 = -2.5*np.log10(s14['F125W']/s14['F160W'])
delta_color = jh_jw - jh_s14[idx]
fig, axes = plt.subplots(3,1,figsize=(8,6), sharex=True, sharey=True)
axes[0].scatter(mag_jw_tot[has_match], delta_mag_aper[has_match], alpha=0.1)
axes[0].set_ylabel(r'$\Delta$mag F160W' + '\nD=0.7" aperture')
axes[1].scatter(mag_jw_tot[has_match], delta_mag_tot[has_match], vmax=1.5, alpha=0.1)
axes[1].set_ylabel(r'$\Delta$mag F160W' + '\n total corrected')
axes[2].scatter(mag_jw_tot[has_match], delta_color[has_match], alpha=0.1)
axes[2].set_ylabel(r'$\Delta$color' + '\nF125W - F160W')
axes[2].set_xlabel('mag, HST F160W')
for ax in axes:
ax.set_xlim(18, 29)
ax.set_ylim(-1,1)
ax.grid()
Photometric redshifts
import eazy.hdf5
if not os.path.exists('templates'):
eazy.symlink_eazy_inputs()
root = f'{field}-fix'
self = eazy.hdf5.initialize_from_hdf5(h5file=root+'.eazypy.h5')
self.fit_phoenix_stars()
zout = utils.read_catalog(root+'.eazypy.zout.fits')
self.cat = utils.read_catalog(root+'_phot_apcorr.fits')
cat = self.cat
Read default param file: /Users/gbrammer/miniconda3/envs/py39jw/lib/python3.9/site-packages/eazy/data/zphot.param.default
CATALOG_FILE is a table
>>> NOBJ = 52427
f090w_tot_1 f090w_etot_1 (363): jwst_nircam_f090w
f105w_tot_1 f105w_etot_1 (202): hst/wfc3/IR/f105w.dat
f110w_tot_1 f110w_etot_1 (241): hst/wfc3/IR/f110w.dat
f115w_tot_1 f115w_etot_1 (364): jwst_nircam_f115w
f115wn_tot_1 f115wn_etot_1 (309): niriss-f115w
f125w_tot_1 f125w_etot_1 (203): hst/wfc3/IR/f125w.dat
f140w_tot_1 f140w_etot_1 (204): hst/wfc3/IR/f140w.dat
f150w_tot_1 f150w_etot_1 (365): jwst_nircam_f150w
f150wn_tot_1 f150wn_etot_1 (310): niriss-f150w
f160w_tot_1 f160w_etot_1 (205): hst/wfc3/IR/f160w.dat
f182m_tot_1 f182m_etot_1 (370): jwst_nircam_f182m
f200w_tot_1 f200w_etot_1 (366): jwst_nircam_f200w
f200wn_tot_1 f200wn_etot_1 (311): niriss-f200w
f210m_tot_1 f210m_etot_1 (371): jwst_nircam_f210m
f277w_tot_1 f277w_etot_1 (375): jwst_nircam_f277w
f335m_tot_1 f335m_etot_1 (381): jwst_nircam_f335m
f356w_tot_1 f356w_etot_1 (376): jwst_nircam_f356w
f410m_tot_1 f410m_etot_1 (383): jwst_nircam_f410m
f430m_tot_1 f430m_etot_1 (384): jwst_nircam_f430m
f435w_tot_1 f435w_etot_1 (233): hst/ACS_update_sep07/wfc_f435w_t81.dat
f444w_tot_1 f444w_etot_1 (377): jwst_nircam_f444w
f460m_tot_1 f460m_etot_1 (385): jwst_nircam_f460m
f475w_tot_1 f475w_etot_1 (234): hst/ACS_update_sep07/wfc_f475w_t81.dat
f480m_tot_1 f480m_etot_1 (386): jwst_nircam_f480m
f606w_tot_1 f606w_etot_1 (236): hst/ACS_update_sep07/wfc_f606w_t81.dat
f606wu_tot_1 f606wu_etot_1 (214): hst/wfc3/UVIS/f606w.dat
f775w_tot_1 f775w_etot_1 (238): hst/ACS_update_sep07/wfc_f775w_t81.dat
f814w_tot_1 f814w_etot_1 (239): hst/ACS_update_sep07/wfc_f814w_t81.dat
f814wu_tot_1 f814wu_etot_1 (217): hst/wfc3/UVIS/f814w.dat
f850lp_tot_1 f850lp_etot_1 (240): hst/ACS_update_sep07/wfc_f850lp_t81.dat
Set sys_err = 0.05 (positive=True)
Read PRIOR_FILE: templates/prior_F160W_TAO.dat
Template grid: templates/sfhz/agn_blue_sfhz_13.param (this may take some time)
TemplateGrid: user-provided tempfilt_data
Process templates: 0.237 s
294it [00:02, 104.84it/s]
h5: read corr_sfhz_13_bin0_av0.01.fits
h5: read corr_sfhz_13_bin0_av0.25.fits
h5: read corr_sfhz_13_bin0_av0.50.fits
h5: read corr_sfhz_13_bin0_av1.00.fits
h5: read corr_sfhz_13_bin1_av0.01.fits
h5: read corr_sfhz_13_bin1_av0.25.fits
h5: read corr_sfhz_13_bin1_av0.50.fits
h5: read corr_sfhz_13_bin1_av1.00.fits
h5: read corr_sfhz_13_bin2_av0.01.fits
h5: read corr_sfhz_13_bin2_av0.50.fits
h5: read corr_sfhz_13_bin2_av1.00.fits
h5: read corr_sfhz_13_bin3_av0.01.fits
h5: read corr_sfhz_13_bin3_av0.50.fits
h5: read fsps_4590.fits
h5: read j0647agn+torus.fits
fit_best: 2.5 s (n_proc=5, NOBJ=51074)
phoenix_templates: ./bt-settl_t400-7000_g4.5.fits
Adjusted zeropoints
The iterated eazy “zeropoint” adjustments are used as a crude PSF-matching correction for the simple aperture photometry catalogs. That is, all photometry is done on the native images and corrected to “total” using a single correction derived in the detection band. The encircled energy (for point sources) will be different for the different instruments / filters, and the eazy zeropoint adjustments are initialized with corrections that would be appropriate to put point sources on a common scale. Corrections to that are then derived based on the photo-z fits to the full catalog.
print('# ix filt f_number zp')
for i in np.argsort(self.lc):
print(f"{i:2} {self.flux_columns[i].split('_')[0]:12} {self.f_numbers[i]:3} {self.zp[i]:.3f}")
# ix filt f_number zp
19 f435w 233 1.067
22 f475w 234 1.091
25 f606wu 214 0.929
24 f606w 236 0.911
26 f775w 238 0.865
27 f814w 239 0.847
28 f814wu 217 1.000
0 f090w 363 0.931
29 f850lp 240 0.875
1 f105w 202 0.932
4 f115wn 309 0.872
2 f110w 241 0.942
3 f115w 364 0.876
5 f125w 203 0.948
6 f140w 204 0.965
8 f150wn 310 0.878
7 f150w 365 0.871
9 f160w 205 0.983
10 f182m 370 0.909
11 f200w 366 0.903
12 f200wn 311 0.882
13 f210m 371 0.936
14 f277w 375 1.000
15 f335m 381 1.052
16 f356w 376 1.077
17 f410m 383 1.114
18 f430m 384 1.138
20 f444w 377 1.148
21 f460m 385 1.175
23 f480m 386 1.205
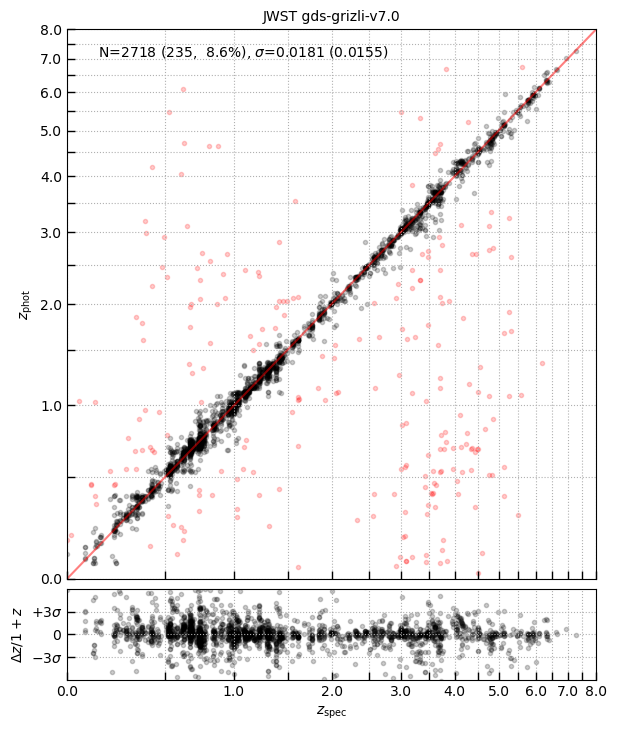
fig = eazy.utils.zphot_zspec(zout['z_phot'][has_match], zout['z_spec'][has_match], zmax=8)
fig.axes[0].set_title(f'JWST {field}')
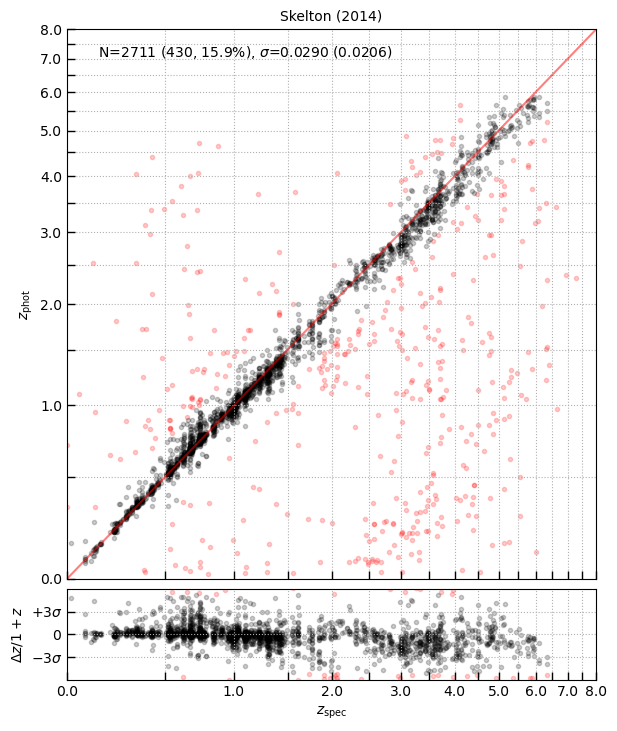
fig = eazy.utils.zphot_zspec(s14['zpk'][idx][has_match], zout['z_spec'][has_match], zmax=8)
fig.axes[0].set_title(f'Skelton (2014)')
Text(0.5, 1.0, 'Skelton (2014)')
Plot some SEDs
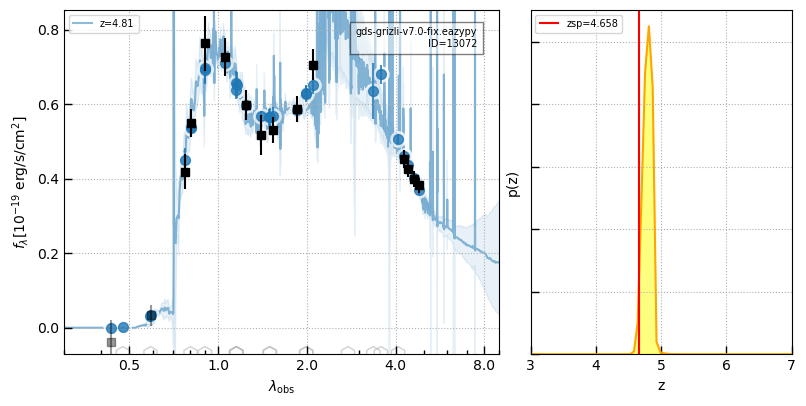
# e.g., GOODS-S-9209 from Carnall et al. https://arxiv.org/pdf/2301.11413.pdf
ra, dec = 53.1082274, -27.8252019
dr = np.sqrt((zout['ra'] - ra)**2 + (zout['dec'] - dec)**2)
self.cat['z_spec'][np.argmin(dr)] = 4.6582
cat_id = zout['id'][np.argmin(dr)]
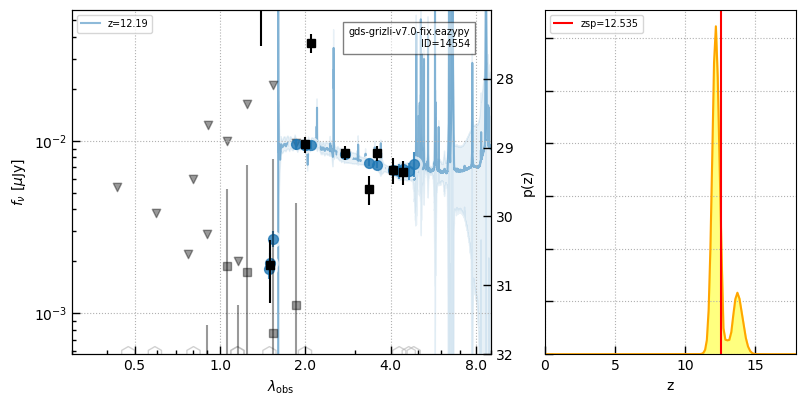
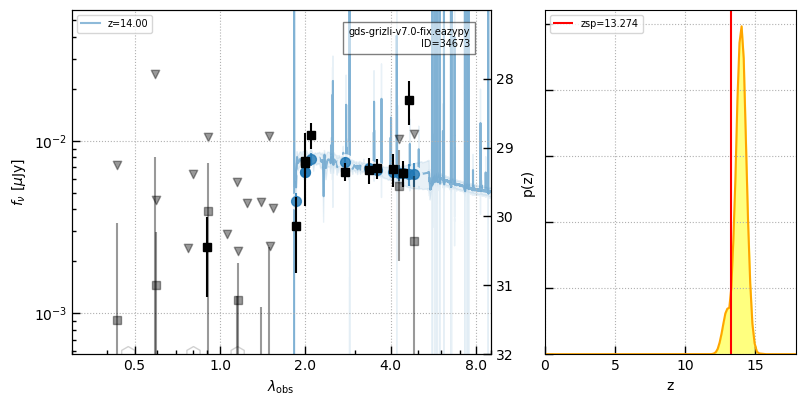
_ = self.show_fit(cat_id, zr=[3,7])
def quick_cutout(resp, sy=2, pl=2, size=1.5, scl=3, filters='f115w-clear,f277w-clear,f444w-clear'):
"""
Make a cutout figure with the grizli cutout server
"""
from PIL import Image
import requests
from io import BytesIO
#rd = ds9.get('pan icrs').replace(' ',',')
if isinstance(resp, int):
id = resp
ix = np.where(self.cat['id'] == id)[0][0]
else:
ix = resp['ix']
id = resp['id']
rd = f"{self.RA[ix]:.6f},{self.DEC[ix]:.6f}"
print(rd)
print(f"https://s3.amazonaws.com/grizli-v2/ClusterTiles/Map/gds/jwst.html?coord={rd}&zoom=6")
url=f"https://grizli-cutout.herokuapp.com/thumb?coord={rd}&all_filters=True&size={size}&scl={scl}&asinh=True&filters={filters}&rgb_scl=1.0,0.95,1.2&pl={pl}"
response = requests.get(url)
img = Image.open(BytesIO(response.content))
img.apply_transparency()
fig, ax = plt.subplots(1,1, figsize=(sy*4, sy+0.2), sharex=True, sharey=True)
data = np.array(img)
black = data.max(axis=2) == 0
for ioff in range(-2,3):
black &= np.roll(black, ioff, axis=0)
for i in range(3):
data[:,:,i][black] = 255
ax.imshow(data, interpolation='Nearest', origin='upper')
#ax.text(0.5, 0.01, 'nrc', color='r', fontsize=8, ha='center',va='bottom',transform=ax.transAxes)
ax.set_xticklabels([])
ax.set_yticklabels([])
ax.axis('off')
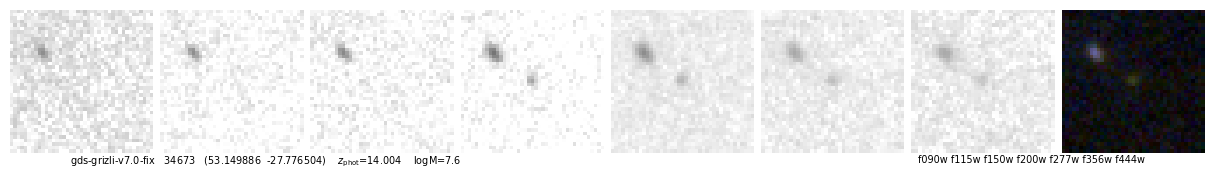
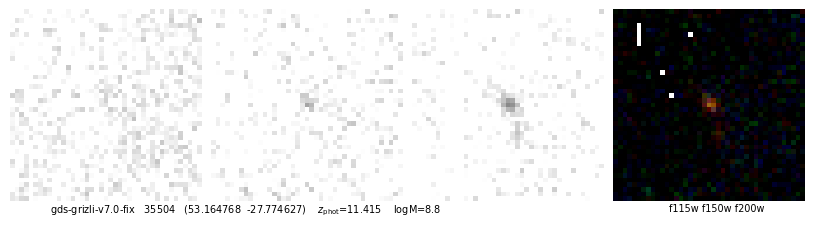
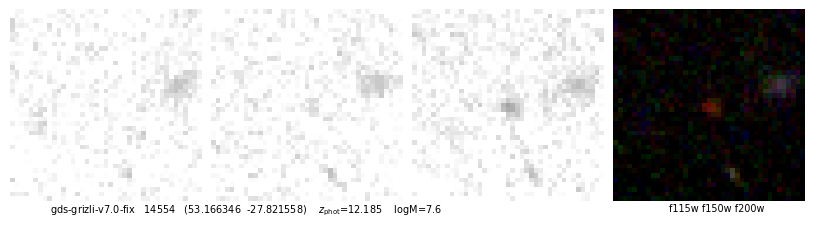
ax.text(0.05, -0.01, f"{root} {self.cat['id'][ix]} ({rd.replace(',',' ')}) z_phot={self.zbest[ix]:.3f} logM={np.log10(zout['mass'][ix]):.1f}".replace('z_phot', r'$z_\mathrm{phot}$'),
ha='left', va='top', transform=ax.transAxes, fontsize=7)
ax.text(0.95, -0.01, filters.replace('-clear','').replace(',', ' '), ha='right', va='top',
fontsize=7, color='k', transform=ax.transAxes)
fig.tight_layout(pad=0.2)
return id, fig, img
if 1:
id, fig, img = quick_cutout(_[1], pl=2, scl=4, sy=3, filters='f775w,f182m-clear,f444w-clear')
# id, fig, img = quick_cutout(_[1], pl=2, scl=5, sy=3)
53.108211,-27.825183
https://s3.amazonaws.com/grizli-v2/ClusterTiles/Map/gds/jwst.html?coord=53.108211,-27.825183&zoom=6

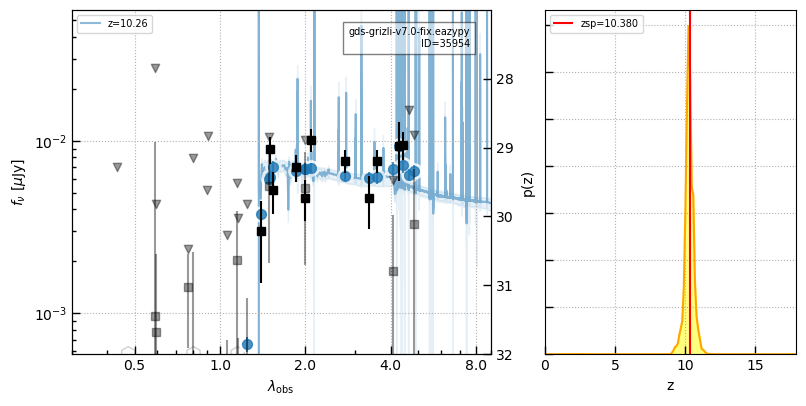
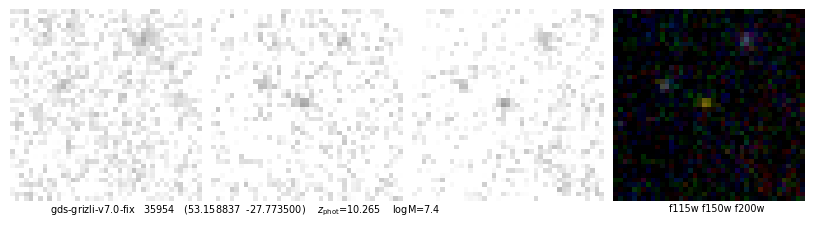
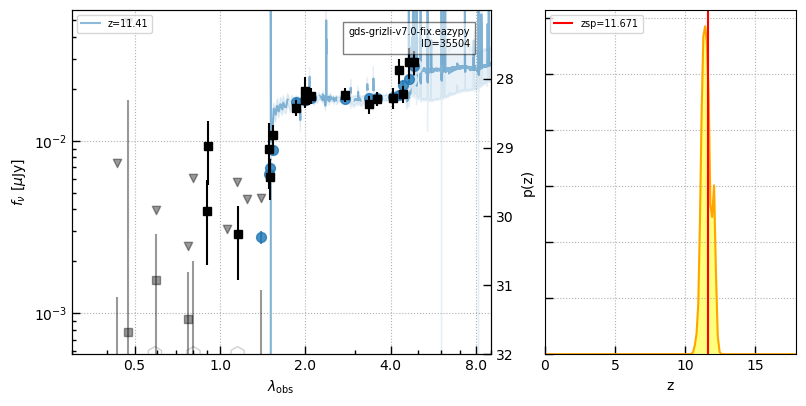
# Curtis Lake - JADES
src = utils.read_catalog('https://raw.githubusercontent.com/dawn-cph/jwst-sources/main/jwst-sources.csv')
idx, dr = zout.match_to_catalog_sky(src)
ecl = src['author'] == 'Emma Curtis-Lake'
ids_jades = zout['id'][idx][ecl]
z_jades = src['zspec'][ecl]
i = -1
self.cat['z_spec'][idx] = src['zspec']
for i in np.argsort(z_jades):
id = ids_jades[i]
_ = self.show_fit(id, show_fnu=True, maglim=(32,27))
# id, fig, img = quick_cutout(_[1], pl=2, scl=8, sy=3, size=1, filters='f090w-clear,f115w-clear,f150w-clear,f200w-clear,f277w-clear,f356w-clear,f444w-clear')
id, fig, img = quick_cutout(_[1], scl=10, size=1, filters='f115w-clear,f150w-clear,f200w-clear')
53.158837,-27.773500
https://s3.amazonaws.com/grizli-v2/ClusterTiles/Map/gds/jwst.html?coord=53.158837,-27.773500&zoom=6
53.164768,-27.774627
https://s3.amazonaws.com/grizli-v2/ClusterTiles/Map/gds/jwst.html?coord=53.164768,-27.774627&zoom=6
53.166346,-27.821558
https://s3.amazonaws.com/grizli-v2/ClusterTiles/Map/gds/jwst.html?coord=53.166346,-27.821558&zoom=6
53.149886,-27.776504
https://s3.amazonaws.com/grizli-v2/ClusterTiles/Map/gds/jwst.html?coord=53.149886,-27.776504&zoom=6
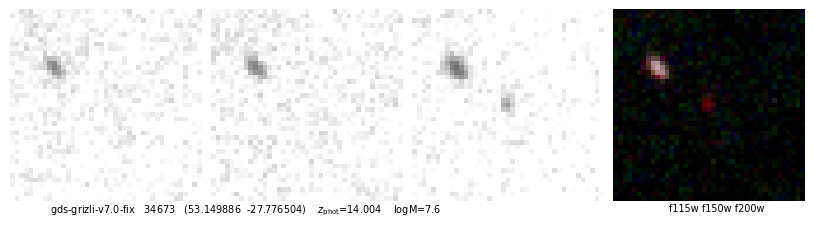
# More filters
id, fig, img = quick_cutout(_[1], pl=2, scl=8, sy=3, size=1, filters='f090w-clear,f115w-clear,f150w-clear,f200w-clear,f277w-clear,f356w-clear,f444w-clear')
53.149886,-27.776504
https://s3.amazonaws.com/grizli-v2/ClusterTiles/Map/gds/jwst.html?coord=53.149886,-27.776504&zoom=6