Gabriel Brammer
(This page is auto-generated from the Jupyter notebook image-data-products.ipynb.)
Here we summarize the files available for imaging datasets, for example in the v7 data release.
%matplotlib inline
import os
import warnings
warnings.filterwarnings('ignore')
import numpy as np
import matplotlib.pyplot as plt
import scipy.ndimage as nd
import astropy.io.fits as pyfits
import astropy.units as u
import sep
import grizli
from grizli import utils
print(f'grizli version: {grizli.__version__}')
BASE_URL = 'https://s3.amazonaws.com/grizli-v2/JwstMosaics/v7/'
grizli version: 1.10.dev3+g341a999
File extensions
Generally, for a given root and filter combination, the following files are available:
{root}-{filter}_drc_sci.fits.gz: Science image{root}-{filter}_drc_wht.fits.gz: Inverse variance weight image (sky + readnoise){root}-{filter}_drc_exp.fits.gz: Exposure-time map{root}-{filter}_wcs.csv: Table summarizing individual exposures that contribute to the mosaic
Notes
- All mosaics are created with the legacy
drizzlepac.adrizzle.do_drizdrizzle implementation that works interchangeably with JWST and HST.grizligenerates WCS for each exposure that follow the SIP-WCS convention and that match the newergwcsJWST wcs at the level of 1e-4 pixels or better
- All NIRCam LW, NIRISS (and HST) mosaics are created with 40 mas pixels
- Most fields have 20 mas pixels for the NIRCam SW images that exacly subsample the LW grid 2x2.
- The very large
primer-cosmosandprimer-udsSW mosaics have 40 mas pixels
- The very large
- All
scimosaics have intensity units of10 nJy / pix, corresponding to an AB magnitude zeropoint 28.9. This has the slightly desirable property that the image pixel values are not too different from unity.- These are not the same as the surface brightness units of the JWST pipeline!
- The
expexposure time images have units of seconds rounded to the nearest integer- Subsampled to 4x4 of the parent mosaic to keep the file sizes small
- The
expimages are created directly from the footprints of the constituent exposures and don’t account for masked pixels within an exposure.
# Example
root = 'smacs0723-grizli-v7.0'
filter = 'f444w-clear'
# Open the files directly from the web
img = {}
print('# File shape')
for ext in ['sci','wht','exp']:
_file = f'{root}-{filter}_drc_{ext}.fits.gz'
img[ext] = pyfits.open(os.path.join(BASE_URL, _file))
print(f'{_file} : {img[ext][0].data.shape}')
# File shape
smacs0723-grizli-v7.0-f444w-clear_drc_sci.fits.gz : (12000, 12000)
smacs0723-grizli-v7.0-f444w-clear_drc_wht.fits.gz : (12000, 12000)
smacs0723-grizli-v7.0-f444w-clear_drc_exp.fits.gz : (3000, 3000)
# Files have a single PrimaryHDU
img['sci'].info()
Filename: /Users/gbrammer/.astropy/cache/download/url/b0380671ce11dec1c5653485f66f705c/contents
No. Name Ver Type Cards Dimensions Format
0 PRIMARY 1 PrimaryHDU 93 (12000, 12000) float32
Primary sci header
img['sci'][0].header
SIMPLE = T / conforms to FITS standard
BITPIX = -32 / array data type
NAXIS = 2 / number of array dimensions
NAXIS1 = 12000
NAXIS2 = 12000
WCSAXES = 2 / Number of coordinate axes
CRPIX1 = 4591.5 / Pixel coordinate of reference point
CRPIX2 = 6515.5 / Pixel coordinate of reference point
CD1_1 = -1.1111111111111E-05 / Coordinate transformation matrix element
CD2_2 = 1.1111111111111E-05 / Coordinate transformation matrix element
CDELT1 = 1.0 / [deg] Coordinate increment at reference point
CDELT2 = 1.0 / [deg] Coordinate increment at reference point
CUNIT1 = 'deg' / Units of coordinate increment and value
CUNIT2 = 'deg' / Units of coordinate increment and value
CTYPE1 = 'RA---TAN' / Right ascension, gnomonic projection
CTYPE2 = 'DEC--TAN' / Declination, gnomonic projection
CRVAL1 = 110.83403 / [deg] Coordinate value at reference point
CRVAL2 = -73.45429 / [deg] Coordinate value at reference point
LONPOLE = 180.0 / [deg] Native longitude of celestial pole
LATPOLE = -73.45429 / [deg] Native latitude of celestial pole
MJDREF = 0.0 / [d] MJD of fiducial time
DATE-OBS= '2022-06-07' / ISO-8601 time of observation
MJD-OBS = 59737.0 / [d] MJD of observation
RADESYS = 'ICRS' / Equatorial coordinate system
CD1_2 = 0.0
CD2_1 = 0.0
DRIZKERN= 'square ' / Drizzle kernel
DRIZPIXF= 0.75 / Drizzle pixfrac
EXPTIME = 15074.42400000001
NDRIZIM = 18
PIXFRAC = 0.75
KERNEL = 'square '
OKBITS = 4 / FLT bits treated as valid
PHOTSCAL= 1.001401962747847 / Scale factor applied
GRIZLIV = '1.8.16.dev12+g86ad0c1' / Grizli code version
WHTTYPE = 'jwst ' / Exposure weighting strategy
RNPERC = 99 / VAR_RNOISE clip percentile for JWST
TELESCOP= 'JWST '
FILTER = 'F444W '
PUPIL = 'CLEAR '
DETECTOR= 'NRCALONG'
INSTRUME= 'NIRCAM '
PHOTFLAM= 1.54184756289340E-22
PHOTPLAM= 44036.71097714713
PHOTFNU = 1E-08
EXPSTART= 59737.22120032604
EXPEND = 59737.23101751157
TIME-OBS= '05:18:31.708'
UPDA_CTX= 'jwst_0995.pmap'
CRDS_CTX= 'jwst_1041.pmap'
R_DISTOR= 'jwst_nircam_distortion_0141.asdf'
R_PHOTOM= 'jwst_nircam_photom_0111.fits'
R_FLAT = 'jwst_nircam_flat_0574.fits'
PHOTMJSR= 0.3925000131130219
PIXAR_SR= 9.31E-14
FLT00001= 'jw02736001001_02105_00001_nrcalong_rate.fits'
WHT00001= 14335.236328125 / Median weight of exposure 1
FLT00002= 'jw02736001001_02105_00001_nrcblong_rate.fits'
WHT00002= 14818.2607421875 / Median weight of exposure 2
FLT00003= 'jw02736001001_02105_00002_nrcalong_rate.fits'
WHT00003= 14245.970703125 / Median weight of exposure 3
FLT00004= 'jw02736001001_02105_00002_nrcblong_rate.fits'
WHT00004= 14709.4404296875 / Median weight of exposure 4
FLT00005= 'jw02736001001_02105_00003_nrcalong_rate.fits'
WHT00005= 14335.7177734375 / Median weight of exposure 5
FLT00006= 'jw02736001001_02105_00003_nrcblong_rate.fits'
WHT00006= 14799.9052734375 / Median weight of exposure 6
FLT00007= 'jw02736001001_02105_00004_nrcalong_rate.fits'
WHT00007= 14333.94921875 / Median weight of exposure 7
FLT00008= 'jw02736001001_02105_00004_nrcblong_rate.fits'
WHT00008= 14851.24609375 / Median weight of exposure 8
FLT00009= 'jw02736001001_02105_00005_nrcalong_rate.fits'
WHT00009= 14321.4130859375 / Median weight of exposure 9
FLT00010= 'jw02736001001_02105_00005_nrcblong_rate.fits'
WHT00010= 14816.029296875 / Median weight of exposure 10
FLT00011= 'jw02736001001_02105_00006_nrcalong_rate.fits'
WHT00011= 14384.9931640625 / Median weight of exposure 11
FLT00012= 'jw02736001001_02105_00006_nrcblong_rate.fits'
WHT00012= 14822.169921875 / Median weight of exposure 12
FLT00013= 'jw02736001001_02105_00007_nrcalong_rate.fits'
WHT00013= 14322.65234375 / Median weight of exposure 13
FLT00014= 'jw02736001001_02105_00007_nrcblong_rate.fits'
WHT00014= 14808.3759765625 / Median weight of exposure 14
FLT00015= 'jw02736001001_02105_00008_nrcalong_rate.fits'
WHT00015= 14282.8134765625 / Median weight of exposure 15
FLT00016= 'jw02736001001_02105_00008_nrcblong_rate.fits'
WHT00016= 14759.8623046875 / Median weight of exposure 16
FLT00017= 'jw02736001001_02105_00009_nrcalong_rate.fits'
WHT00017= 14353.7353515625 / Median weight of exposure 17
FLT00018= 'jw02736001001_02105_00009_nrcblong_rate.fits'
WHT00018= 14783.2705078125 / Median weight of exposure 18
OPHOTFNU= 9.30775449348276E-08 / Original PHOTFNU before scaling
BUNIT = '10.0*nanoJansky'
# Images have units of 10 nJy / pix
for k in ('FILTER','PHOTFNU','PHOTPLAM','BUNIT'):
print(f"{k:>8}: {img['sci'][0].header[k]}")
FILTER: F444W
PHOTFNU: 1e-08
PHOTPLAM: 44036.71097714713
BUNIT: 10.0*nanoJansky
Primary exp header
img['exp'][0].header
SIMPLE = T / conforms to FITS standard
BITPIX = 32 / array data type
NAXIS = 2 / number of array dimensions
NAXIS1 = 3000
NAXIS2 = 3000
WCSAXES = 2 / Number of coordinate axes
CRPIX1 = 1147.875 / Pixel coordinate of reference point
CRPIX2 = 1628.875 / Pixel coordinate of reference point
CD1_1 = -4.4444444444444E-05 / Coordinate transformation matrix element
CD2_2 = 4.4444444444444E-05 / Coordinate transformation matrix element
CDELT1 = 1.0 / [deg] Coordinate increment at reference point
CDELT2 = 1.0 / [deg] Coordinate increment at reference point
CUNIT1 = 'deg' / Units of coordinate increment and value
CUNIT2 = 'deg' / Units of coordinate increment and value
CTYPE1 = 'RA---TAN' / Right ascension, gnomonic projection
CTYPE2 = 'DEC--TAN' / Declination, gnomonic projection
CRVAL1 = 110.83403 / [deg] Coordinate value at reference point
CRVAL2 = -73.45429 / [deg] Coordinate value at reference point
LONPOLE = 180.0 / [deg] Native longitude of celestial pole
LATPOLE = -73.45429 / [deg] Native latitude of celestial pole
MJDREF = 0.0 / [d] MJD of fiducial time
DATE-OBS= '2022-06-07' / ISO-8601 time of observation
MJD-OBS = 59737.0 / [d] MJD of observation
RADESYS = 'ICRS' / Equatorial coordinate system
CD1_2 = 0.0
CD2_1 = 0.0
DRIZKERN= 'square ' / Drizzle kernel
DRIZPIXF= 0.75 / Drizzle pixfrac
EXPTIME = 15074.42400000001
NDRIZIM = 18
PIXFRAC = 0.75
KERNEL = 'square '
OKBITS = 4 / FLT bits treated as valid
PHOTSCAL= 1.001401962747847 / Scale factor applied
GRIZLIV = '1.8.16.dev12+g86ad0c1' / Grizli code version
WHTTYPE = 'jwst ' / Exposure weighting strategy
RNPERC = 99 / VAR_RNOISE clip percentile for JWST
TELESCOP= 'JWST '
FILTER = 'F444W '
PUPIL = 'CLEAR '
DETECTOR= 'NRCALONG'
INSTRUME= 'NIRCAM '
PHOTFLAM= 1.54184756289340E-22
PHOTPLAM= 44036.71097714713
PHOTFNU = 1E-08
EXPSTART= 59737.22120032604
EXPEND = 59737.23101751157
TIME-OBS= '05:18:31.708'
UPDA_CTX= 'jwst_0995.pmap'
CRDS_CTX= 'jwst_1041.pmap'
R_DISTOR= 'jwst_nircam_distortion_0141.asdf'
R_PHOTOM= 'jwst_nircam_photom_0111.fits'
R_FLAT = 'jwst_nircam_flat_0574.fits'
PHOTMJSR= 0.3925000131130219
PIXAR_SR= 9.31E-14
FLT00001= 'jw02736001001_02105_00001_nrcalong_rate.fits'
WHT00001= 14335.236328125 / Median weight of exposure 1
FLT00002= 'jw02736001001_02105_00001_nrcblong_rate.fits'
WHT00002= 14818.2607421875 / Median weight of exposure 2
FLT00003= 'jw02736001001_02105_00002_nrcalong_rate.fits'
WHT00003= 14245.970703125 / Median weight of exposure 3
FLT00004= 'jw02736001001_02105_00002_nrcblong_rate.fits'
WHT00004= 14709.4404296875 / Median weight of exposure 4
FLT00005= 'jw02736001001_02105_00003_nrcalong_rate.fits'
WHT00005= 14335.7177734375 / Median weight of exposure 5
FLT00006= 'jw02736001001_02105_00003_nrcblong_rate.fits'
WHT00006= 14799.9052734375 / Median weight of exposure 6
FLT00007= 'jw02736001001_02105_00004_nrcalong_rate.fits'
WHT00007= 14333.94921875 / Median weight of exposure 7
FLT00008= 'jw02736001001_02105_00004_nrcblong_rate.fits'
WHT00008= 14851.24609375 / Median weight of exposure 8
FLT00009= 'jw02736001001_02105_00005_nrcalong_rate.fits'
WHT00009= 14321.4130859375 / Median weight of exposure 9
FLT00010= 'jw02736001001_02105_00005_nrcblong_rate.fits'
WHT00010= 14816.029296875 / Median weight of exposure 10
FLT00011= 'jw02736001001_02105_00006_nrcalong_rate.fits'
WHT00011= 14384.9931640625 / Median weight of exposure 11
FLT00012= 'jw02736001001_02105_00006_nrcblong_rate.fits'
WHT00012= 14822.169921875 / Median weight of exposure 12
FLT00013= 'jw02736001001_02105_00007_nrcalong_rate.fits'
WHT00013= 14322.65234375 / Median weight of exposure 13
FLT00014= 'jw02736001001_02105_00007_nrcblong_rate.fits'
WHT00014= 14808.3759765625 / Median weight of exposure 14
FLT00015= 'jw02736001001_02105_00008_nrcalong_rate.fits'
WHT00015= 14282.8134765625 / Median weight of exposure 15
FLT00016= 'jw02736001001_02105_00008_nrcblong_rate.fits'
WHT00016= 14759.8623046875 / Median weight of exposure 16
FLT00017= 'jw02736001001_02105_00009_nrcalong_rate.fits'
WHT00017= 14353.7353515625 / Median weight of exposure 17
FLT00018= 'jw02736001001_02105_00009_nrcblong_rate.fits'
WHT00018= 14783.2705078125 / Median weight of exposure 18
OPHOTFNU= 9.30775449348276E-08 / Original PHOTFNU before scaling
BUNIT = 'second '
SAMPLE = 4 / Sampling factor
NXORIG = 12000
NYORIG = 12000
MOSPSCL = 0.03999999999999958 / Mosaic pixel scale arcsec
ORIGPSCL= 0.0629361212228063 / Original detector pixel scale arcsec
DNTOEPS = 58.62417855098175 / Inverse flux conversion back to e per second
BSCALE = 1
BZERO = 2147483648
WCS log
The wcs.csv files contain the full SIP header of each exposure that contributes to the mosaic, along with some epoch information.
_file = f'{root}-{filter}_wcs.csv'
wcs = utils.read_catalog(os.path.join(BASE_URL, _file))
print(wcs.colnames)
['file', 'ext', 'exptime', 'wcsaxes', 'crpix1', 'crpix2', 'cd1_1', 'cd1_2', 'cd2_1', 'cd2_2', 'cdelt1', 'cdelt2', 'cunit1', 'cunit2', 'ctype1', 'ctype2', 'crval1', 'crval2', 'lonpole', 'latpole', 'wcsname', 'mjdref', 'date-beg', 'mjd-beg', 'date-avg', 'mjd-avg', 'date-end', 'mjd-end', 'xposure', 'telapse', 'obsgeo-x', 'obsgeo-y', 'obsgeo-z', 'radesys', 'velosys', 'a_order', 'a_0_2', 'a_0_3', 'a_0_4', 'a_0_5', 'a_1_1', 'a_1_2', 'a_1_3', 'a_1_4', 'a_2_0', 'a_2_1', 'a_2_2', 'a_2_3', 'a_3_0', 'a_3_1', 'a_3_2', 'a_4_0', 'a_4_1', 'a_5_0', 'b_order', 'b_0_2', 'b_0_3', 'b_0_4', 'b_0_5', 'b_1_1', 'b_1_2', 'b_1_3', 'b_1_4', 'b_2_0', 'b_2_1', 'b_2_2', 'b_2_3', 'b_3_0', 'b_3_1', 'b_3_2', 'b_4_0', 'b_4_1', 'b_5_0', 'naxis', 'naxis1', 'naxis2', 'sipcrpx1', 'sipcrpx2']
# First few lines
wcs['file','ext','exptime','mjd-avg','date-avg','crpix1','crpix2','crval1','crval2'][:4]
| file | ext | exptime | mjd-avg | date-avg | crpix1 | crpix2 | crval1 | crval2 |
|---|---|---|---|---|---|---|---|---|
| str44 | int64 | float64 | float64 | str23 | float64 | float64 | float64 | float64 |
| jw02736001001_02105_00001_nrcalong_rate.fits | 1 | 837.468 | 59737.226108919 | 2022-06-07T05:25:35.811 | 1024.647 | 1024.66 | 110.68612332528 | -73.481395298838 |
| jw02736001001_02105_00001_nrcblong_rate.fits | 1 | 837.468 | 59737.226105215 | 2022-06-07T05:25:35.491 | 1024.489 | 1024.662 | 110.8276309259 | -73.453986741388 |
| jw02736001001_02105_00002_nrcalong_rate.fits | 1 | 837.468 | 59737.236795574 | 2022-06-07T05:40:59.138 | 1024.647 | 1024.66 | 110.68291892635 | -73.47999884627 |
| jw02736001001_02105_00002_nrcblong_rate.fits | 1 | 837.468 | 59737.23679187 | 2022-06-07T05:40:58.818 | 1024.489 | 1024.662 | 110.82443032455 | -73.452590822835 |
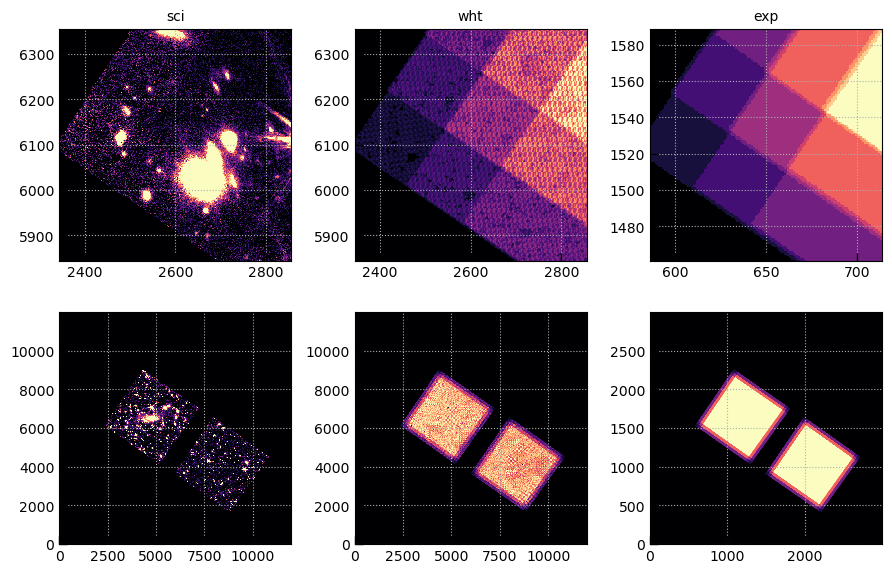
Compare the WHT and EXP images
exts = ['sci','wht','exp']
fig, axes = plt.subplots(2,len(exts),figsize=(3*len(exts),6))
for j, ext in enumerate(exts):
msk = img[ext][0].data != 0
wmax = np.nanpercentile(img[ext][0].data[msk], 95)
for i in [0,1]:
axes[i][j].imshow(img[ext][0].data, vmin=0, vmax=wmax, origin='lower', cmap='magma')
axes[i][j].grid()
if i == 0:
axes[i][j].set_title(ext)
xy = 2600, 6100, 256
for j, p in enumerate([0,0,1]):
axes[0][j].set_xlim(*(xy[0] + np.array([-1,1])*xy[2])/4**p)
axes[0][j].set_ylim(*(xy[1] + np.array([-1,1])*xy[2])/4**p)
fig.tight_layout(pad=1)
Make a full variance image including the Poisson component from the sources themselves
The mosaics are created by weighting each input exposure by a factor like 1/wht = VAR_RNOISE + median(VAR_POISSON) from the JWST exposure files. The first term incorporates pixel-to-pixel variations resulting from pixels where some fraction of the reads may have been masked as saturated or affected by cosmic rays. The second term effectively provides the noise from the sky background, but without including the Poisson term for individual sources.
Certain applications like photometry or morphology fitting with galfit may require a variance / sigma image that includes the poisson term from the individual sources. This can be generated from the exp maps as shown below.
# Grow the exposure map to the original frame
full_exp = np.zeros(img['sci'][0].data.shape, dtype=int)
full_exp[2::4,2::4] += img['exp'][0].data*1
full_exp = nd.maximum_filter(full_exp, 4)
img['Full exp'] = pyfits.HDUList([pyfits.PrimaryHDU(data=full_exp)])
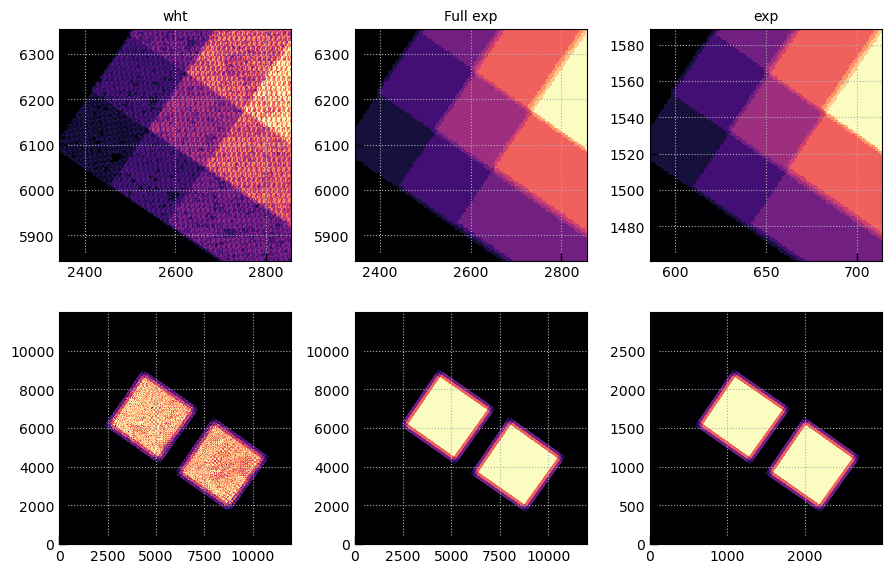
# Show the full exposure map
exts = ['wht','Full exp','exp']
fig, axes = plt.subplots(2,len(exts),figsize=(3*len(exts),6))
for j, ext in enumerate(exts):
msk = img[ext][0].data != 0
wmax = np.nanpercentile(img[ext][0].data[msk], 95)
for i in [0,1]:
axes[i][j].imshow(img[ext][0].data, vmin=0, vmax=wmax, origin='lower', cmap='magma')
axes[i][j].grid()
if i == 0:
axes[i][j].set_title(ext)
xy = 2600, 6100, 256
for j, p in enumerate([0,0,1]):
axes[0][j].set_xlim(*(xy[0] + np.array([-1,1])*xy[2])/4**p)
axes[0][j].set_ylim(*(xy[1] + np.array([-1,1])*xy[2])/4**p)
fig.tight_layout(pad=1)
Effective “gain”
To convert the Poisson variance associated with the sci image, write down the multiplicative factors that had been applied to the original count-rate data in the pipeline rate files. PHOTMJSR is the original flux calibration to units of “MJy/sr” and PHOTSCL is any (small) additional photometric term that was included by grizli. The PHOTFNU / OPHOTFNU term accounts for any final scaling of the output mosaic and the ratio of the original and mosaic pixel areas.
# Scale factors
phot_scale = 1 / (PHOTMJSR * PHOTSCAL) * PHOTFNU / OPHOTFNU
# Effective gain e-/DN, including exposure time
effective_gain = phot_scale * exposure_time_map
# Variance in electrons = counts in electrons
var_poisson_elec = sci * effective_gain
# Variance in mosaic DN
var_poisson_dn = var_poisson_elec / effective_gain**2 = sci / effective_gain
header = img['exp'][0].header
# Multiplicative factors that have been applied since the original count-rate images
phot_scale = 1.
for k in ['PHOTMJSR','PHOTSCAL']:
print(f'{k} {header[k]:.3f}')
phot_scale /= header[k]
# Unit and pixel area scale factors
if 'OPHOTFNU' in header:
phot_scale *= header['PHOTFNU'] / header['OPHOTFNU']
# "effective_gain" = electrons per DN of the mosaic
effective_gain = (phot_scale * full_exp)
# Poisson variance in mosaic DN
var_poisson_dn = np.maximum(img['sci'][0].data, 0) / effective_gain
# Original variance from the `wht` image = RNOISE + BACKGROUND
var_wht = 1/img['wht'][0].data
# New total variance
var_total = var_wht + var_poisson_dn
full_wht = 1 / var_total
# Null weights
full_wht[var_total <= 0] = 0
img['Full wht'] = pyfits.HDUList([pyfits.PrimaryHDU(data=full_wht, header=img['wht'][0].header)])
PHOTMJSR 0.393
PHOTSCAL 1.001
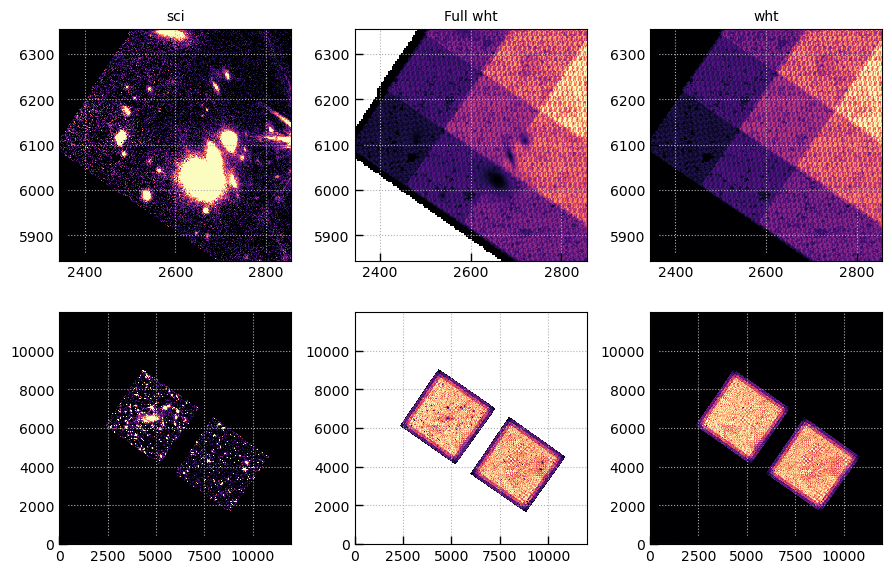
# Compare science and weight arrays, where you can now
# "see" the sources in the Full weight array
exts = ['sci','Full wht','wht']
fig, axes = plt.subplots(2,len(exts),figsize=(3*len(exts),6))
for j, ext in enumerate(exts):
msk = img[ext][0].data != 0
wmax = np.nanpercentile(img[ext][0].data[msk], 95)
for i in [0,1]:
axes[i][j].imshow(img[ext][0].data, vmin=0, vmax=wmax, origin='lower', cmap='magma')
axes[i][j].grid()
if i == 0:
axes[i][j].set_title(ext)
xy = 2600, 6100, 256
for j, p in enumerate([0,0,0]):
axes[0][j].set_xlim(*(xy[0] + np.array([-1,1])*xy[2])/4**p)
axes[0][j].set_ylim(*(xy[1] + np.array([-1,1])*xy[2])/4**p)
fig.tight_layout(pad=1)