Gabriel Brammer
spectroscopy nirspec release catalog
(This page is auto-generated from the Jupyter notebook nirspec-merged-table-v4.ipynb.)
Demo of full merged table of NIRSpec spectra reduced with msaexp. The merged columns are taken from the database tables
nirspec_extractions- Basic spectrum parameters (grating, mask, exposure time, etc.)nirspec_redshifts- Redshift fit results, emission line fluxesnirspec_redshifts_manual- Grades and comments from visual inspectionnirspec_integrated- Observed- and rest-frame filters integrated through the spectra at the derived redshiftgrizli_photometry- Photometry and some eazy outputs of the nearest counterpart in the DJA/grizli photometric catalogs
The public spectra are shown in a large overview table at public_prelim_v4.2.html.
5 September 2025
The notebook has been updated to use a new version v4.4 of the merged table.
- Includes PRISM spectra from the CANUCS GTO program, along with a number of additional public datasets
- Merged files of the 1D spectra in all gratings are now available, with some grating examples added at the end of the demo.
- The summary table and tables of compiled spectra now include all available spectra, whether or not they had been processed with the redshift fit algorithm.
- Companion overview table at nirspec_public_v4.4.html. (Now has the same length as the summary table here.)
- All files provided at the static DOI 10.5281/zenodo.1547235. See README.md at the zenodo page for more information on the release contents.
README
This repository is a snapshot of the public JWST NIRSpec spectra processed with the msaexp pipeline. Please refer to and cite de Graaff et al. (2024) and Heintz et al. (2025) for the main presentation of the msaexp pipeline.
This release corresponds to the v4 version of the spectral extractions, which significantly extends the wavelength range of the extracted spectra to regions that may suffer contamination of overlapping spectral orders. The sensitivity of the higher orders is strongly weighted toward the blue side of the spectrum for all gratings, so, in practice, relatively red galaxies often suffer relatively minor order contamination.
Please refer to and cite this DOI 10.5281/zenodo.1547235 and Valentino et al. (2025) when using this specific data release and the v4 spectra for a presentation of the extended extractions. For more information and updates, please refer to the DJA Blog Post: https://dawn-cph.github.io/dja/blog/2025/05/01/nirspec-merged-table-v4/
Data Content
This release provides version 4 of the NIRSpec Merged Table, a comprehensive catalog of uniformly reduced and analyzed JWST/NIRSpec spectra. The data have been processed using the msaexp and grizli pipelines and are publicly available through the DAWN JWST Archive (DJA). The catalog integrates multiple data products, including spectral extractions, redshift measurements, emission line fluxes, and photometric associations.
The merged table consolidates information from several database tables:
nirspec_extractions: Basic spectrum parameters (e.g., grating, mask, exposure time).nirspec_redshifts: Redshift fit results and emission line fluxes.nirspec_redshifts_manual: Grades and comments from visual inspection.nirspec_integrated: Observed- and rest-frame filters integrated through the spectra at the derived redshift.grizli_photometry: Photometry and some EAZY outputs of the nearest counterpart in the DJA/grizli photometric catalogs.
The catalog includes 80,367 entries, each corresponding to a unique NIRSpec spectrum. Each entry contains metadata such as source ID, coordinates, grating/filter configuration, exposure time, redshift estimates, emission line measurements, and photometric associations.
| N | Grating-Filter | Concatenated 1D spectrum file |
|---|---|---|
| 113 | G140H-F070LP | dja_msaexp_emission_lines_v4.4.g140h-f070lp_spectra.fits |
| 684 | G140H-F100LP | dja_msaexp_emission_lines_v4.4.g140h-f100lp_spectra.fits |
| 5851 | G140M-F070LP | dja_msaexp_emission_lines_v4.4.g140m-f070lp_spectra.fits |
| 2165 | G140M-F100LP | dja_msaexp_emission_lines_v4.4.g140m-f100lp_spectra.fits |
| 6179 | G235H-F170LP | dja_msaexp_emission_lines_v4.4.g235h-f170lp_spectra.fits |
| 8000 | G235M-F170LP | dja_msaexp_emission_lines_v4.4.g235m-f170lp_spectra.fits |
| 8820 | G395H-F290LP | dja_msaexp_emission_lines_v4.4.g395h-f290lp_spectra.fits |
| 13606 | G395M-F290LP | dja_msaexp_emission_lines_v4.4.g395m-f290lp_spectra.fits |
| 34949 | PRISM-CLEAR | dja_msaexp_emission_lines_v4.4.prism_spectra.fits |
Usage Notes
- Data Format: The main catalog is provided in compressed CSV format. Column descriptions, including units and formats, are detailed in the accompanying columns CSV file.
The redshift and line fluxes are derived with the functions msaexp.fit_redshift and msaexp.plot_spectrum. The continuum is modelled as a combination of splines, whose coefficient are listed in the table. Emission lines are superimposed as Gaussian profiles, smoothed by the instrumental resolution (increased by a factor of 1.3x compared with the nominal line spread functions in the JWST User Documentation, JDox, see de Graaff+2025) and a fixed line velocity width of 100 km/s. Equivalent widths in angstrom in the observed frame are also reported. A dictionary with the available lines and their rest-frame wavelengths in vacuum can be generated with grizli.utils.get_line_wavelengths(). The fit is performed with a least square template method. The uncertainties are rescaled with a polynomial curve prior to fitting such that (flux - model) / (err * scl) residuals are N(0,1) using msaexp.spectrum.calc_uncertainty_scale. A systematic uncertainty floor of 2% is introduced. The robustness of the redshift solution is flagged with visual inspection according to the following scheme:
- Grade 3: Robust redshift from one or more emission absorption features
- Grade 2: Ambiguous continuum features, perhaps only one line or low confidence lines
- Grade 1: No clear features in the spectrum to constrain the redshift
- Grade 0: Spectrum suffers some data quality issue and should
- Grade -1: Fit not performed or graded
If multiple spectra of the same sources are available, a common best redshift solution is stored in the z_best column.
-
Spectral Data: Each entry includes links to the corresponding 2D spectra in fits and png format, which can be accessed through the DJA interface or the public spectra overview page at: nirspec_public_v4.4.html The 1D spectra in the fits tables in this release are in the format generated by
msaexp. The columns are described in dja_msaexp_emission_lines_v4.4.columns.csv. -
Some masks that provide spectra of predominantly local sources in the MW or in nearby galaxies were not processed through the redshift fitting procedure. The general spectrum metadata is included in the summary table and the spectra are include in the compiled tables, but the columns of quantities derived from the redshift fit (e.g., emission line fluxes) are not populated.
-
If existing, a comparison with the previous v3 version the spectra generated with msaexp is available at the same link on the DJA interface.
-
Photometric Associations: Photometric data are matched to the nearest counterparts in the DJA/grizli catalogs, providing additional context for each spectroscopic observation. The latest public versions of the DJA/grizli catalogs can are described here: https://dawn-cph.github.io/dja/imaging/v7/ The latest
v7*version of the imaging mosaics and catalogs can be retrieved here: https://s3.amazonaws.com/grizli-v2/JwstMosaics/v7/index.html A description of the data reduction and catalog creation is available on the DJA interface and in Valentino+2023.
Caveats
- An effective extended-source path-loss correction for light outside of the slitlet for each source using the a priori position within the shutter and assuming an azimuthally symmetric Gaussian profile is applied to each spectrum in this release (de Graaff et al. 2025, Section 3). However, the spectra are not rescaled to match the observed photometry in the “phot_” columns.
- The flux calibration is derived from calibration, monitoring, and scientific programs (Valentino et al. 2025). However, residual features in spectra due to an imperfect cross-calibration of different overlapping orders are still present in the released spectra.
- Examples of second order corrections are presented in Ito et al. (2025), where medium-resolution grating spectra beyond their nominal coverage deviate from the prism counterpart (Figure C.1 in appendix in Ito et al. 2025). Moreover, a significant downturn is present in prism spectra at wavelengths longer than 5.2 µm. These second order corrections are largely mitigated cross-calibrating all the available spectra and photometry by means of simple low-order polynomial corrections, for example available in spectrophotometric modeling codes. Future calibration programs dedicated to reconstruction of the sensitivity curves in the extended spectra at different location of the MSA will allow for refined absolute calibrations.
Software
The data processing and analysis utilized the following software packages:
- msaexp:
v0.9.8.dev3+ge0e3f39.d20250429 - grizli:
v1.12.12.dev5+g5896d62.d20250426 - eazy-py:
v0.8.5
Citations
We encourage users to refer to the original works describing the datasets collected in this release. Relevant references, compiled to the best of our knowledge, are available here: https://dawn-cph.github.io/dja/spectroscopy/nirspec/. Please file an Issue on the DJA website repository if you would prefer a different citation for your own dataset.
Works that make use of the products of the DJA should cite this DOI and relevant articles describing them:
1) Msaexp pipeline and methods, v3 and previous spectroscopic compilation releases:
- de Graaff, A., Brammer, G., Weibel, A., et al., “RUBIES: a complete census of the bright and red distant Universe with JWST/NIRSpec”, A&A, 697, 189 (2025)
- Heintz, K. E., Brammer, G., Watson, D., et al., “The JWST-PRIMAL archival survey: A JWST/NIRSpec reference sample for the physical properties and Lyman-α absorption and emission of ∼600 galaxies at z = 5.0−13.4”, A&A, 693, 60 (2025)
- Brammer G., “msaexp: NIRSpec analyis tools”, 10.5281/zenodo.7299500 (2022)
2) Spectroscopic release v4 and extended spectra:
- Valentino, F., Heintz, K. E., Brammer, G. et al., “Gas outflows in two recently quenched galaxies at z = 4 and 7”, A&A, 699, 358 (2025)
- Pollock, C., Gottumukkala, R., Heintz, K. E. et al., “Novel z~10 auroral line measurements extend the gradual offset of the FMR deep into the first Gyr of cosmic time “, arXiv:2506.15779 (2025)
3) Grizli pipeline:
- Brammer G., “grizli”, https://zenodo.org/records/8370018 (2023)
4) Imaging release:
- Valentino, F., Brammer, G., Gould, K. M. L. et al., “An Atlas of Color-selected Quiescent Galaxies at z > 3 in Public JWST Fields”, ApJ, 947, 20 (2023)
Demo
# Install dependencies, e.g., on Google Colab
try:
import msaexp
except ImportError:
! pip install msaexp
! pip install git+https://github.com/karllark/dust_attenuation.git
import eazy
eazy.fetch_eazy_photoz()
%matplotlib inline
import os
import yaml
import numpy as np
import matplotlib.pyplot as plt
from tqdm import tqdm
import warnings
warnings.filterwarnings('ignore')
from scipy.spatial import cKDTree
import astropy.io.fits as pyfits
from astropy.utils.data import download_file
from astropy.cosmology import WMAP9
import astropy.units as u
import grizli
import grizli.catalog
from grizli import utils
import eazy
import msaexp
CACHE_DOWNLOADS = True
print(f'grizli version: {grizli.__version__}')
print(f'eazy-py version: {eazy.__version__}')
print(f'msaexp version: {msaexp.__version__}')
grizli version: 1.13.2.dev1+g1439c13c5.d20250908
eazy-py version: 0.8.5
msaexp version: 0.9.12.dev20+g2d24ffa54.d20250903
Read the table
# Full table
version = "v4.0" # Original notebook release
version = "v4.3" # Updated August 11, 2025. Includes CANUCS and some other additional masks.
version = "v4.4" # Updated September 5, 2025. Include all public spectra even without redshift / line fits
URL_PREFIX = "https://s3.amazonaws.com/msaexp-nirspec/extractions"
# Use the Zenodo release
if version == "v4.4":
URL_PREFIX = "https://zenodo.org/records/15472354/files/"
table_url = f"{URL_PREFIX}/dja_msaexp_emission_lines_{version}.csv.gz"
tab = utils.read_catalog(download_file(table_url, cache=CACHE_DOWNLOADS), format='csv')
Column descriptions
columns_url = f"{URL_PREFIX}/dja_msaexp_emission_lines_{version}.columns.csv"
tab_columns = utils.read_catalog(download_file(columns_url, cache=CACHE_DOWNLOADS), format='csv')
# Set column metadata
for row in tab_columns:
c = row['column']
if row['unit'] != '--':
tab[c].unit = row['unit']
if row['format'] != '--':
tab[c].format = row['format']
if row['description'] != '--':
tab[c].description = row['description']
tab.info()
<GTable length=80367>
name dtype unit format description class n_bad
------------------- ------- ------ ------ ---------------------------------------------------------------------------------------- ------------ -----
file str57 DJA filename Column 0
srcid int64 Source ID from APT plan Column 0
ra float64 deg .8f RA from APT plan Column 0
dec float64 deg .8f Dec from APT plan Column 0
grating str5 NIRSpec grating Column 0
filter str6 Blocking filter Column 0
effexptm float64 Effective exposure time of each exposure Column 0
nfiles int64 Number of files combined in final spectrum Column 0
dataset str72 Filename of first exposure Column 0
msamet str25 MSA metadata file Column 0
msaid int64 MSA metadata ID Column 0
msacnf int64 MSA metadata config Column 0
dithn int64 Dither number Column 0
slitid int64 MSA plan slit ID Column 0
root str24 DJA program + mask rootname Column 0
npix int64 Number of pixels in the 2D spectrum Column 0
ndet int64 Number of detectors contributing to output Column 0
wmin float64 micron Minimum wavelength of the combined spectrum Column 0
wmax float64 micron Maximum wavelength of the combined spectrum Column 0
wmaxsn float64 micron Wavelength of maximum signal to noise Column 0
sn10 float64 10th percentile SN Column 0
flux10 float64 Flux at sn10 Column 0
err10 float64 Uncertainty at sn50 Column 0
sn50 float64 50th percentile SN Column 0
flux50 float64 Flux at sn50 Column 0
err50 float64 Uncertainty at sn90 Column 0
sn90 float64 90th percentile SN Column 0
flux90 float64 Flux at sn90 Column 0
err90 float64 Uncertainty at sn90 Column 0
xstart int64 Starting detector x coordinate of 2D cutout Column 0
ystart int64 Starting detector y coordinate of 2D cutout Column 0
xsize int64 x size of 2D cutout Column 0
ysize int64 y size of 2D cutout Column 0
slit_pa float64 Estimated PA of the slitlet Column 0
pa_v3 float64 Estimated PA of the spacecraft V3 axis Column 0
srcypix float64 Location of the source in the 2D spectrum Column 0
profcen float64 Profile offset relative to the expected center Column 0
profsig float64 Derived profile width in pixels added to PSF Column 0
ctime float64 UNIX time when file was generated Column 0
version str30 MSAEXP code version Column 0
exptime float64 Estimated total exposure time Column 0
contchi2 float64 Chi2 of the spline continuum fit MaskedColumn 18242
dof int64 Total number of pixels in the redshift + line fit MaskedColumn 18242
fullchi2 float64 Chi2 of the full continuum + line fit MaskedColumn 18242
line_ariii_7138 float64 Line flux of ariii_7138 1e-20 erg/s/cm2 MaskedColumn 35864
line_ariii_7138_err float64 MaskedColumn 35868
line_ariii_7753 float64 Line flux of ariii_7753 1e-20 erg/s/cm2 MaskedColumn 37381
line_ariii_7753_err float64 MaskedColumn 37382
line_bra float64 Line flux of bra 1e-20 erg/s/cm2 MaskedColumn 77651
line_bra_err float64 MaskedColumn 77651
line_brb float64 Line flux of brb 1e-20 erg/s/cm2 MaskedColumn 71170
line_brb_err float64 MaskedColumn 71170
line_brd float64 Line flux of brd 1e-20 erg/s/cm2 MaskedColumn 64722
line_brd_err float64 MaskedColumn 64722
line_brg float64 Line flux of brg 1e-20 erg/s/cm2 MaskedColumn 67432
line_brg_err float64 MaskedColumn 67432
line_hb float64 Line flux of hb 1e-20 erg/s/cm2 MaskedColumn 36409
line_hb_err float64 MaskedColumn 36409
line_hd float64 Line flux of hd 1e-20 erg/s/cm2 MaskedColumn 40270
line_hd_err float64 MaskedColumn 40270
line_hei_1083 float64 Line flux of hei_1083 1e-20 erg/s/cm2 MaskedColumn 46220
line_hei_1083_err float64 MaskedColumn 46223
line_hei_3889 float64 Line flux of hei_3889 1e-20 erg/s/cm2 MaskedColumn 51316
line_hei_3889_err float64 MaskedColumn 51319
line_hei_5877 float64 Line flux of hei_5877 1e-20 erg/s/cm2 MaskedColumn 34474
line_hei_5877_err float64 MaskedColumn 34474
line_hei_7065 float64 Line flux of hei_7065 1e-20 erg/s/cm2 MaskedColumn 35664
line_hei_7065_err float64 MaskedColumn 35669
line_hei_8446 float64 Line flux of hei_8446 1e-20 erg/s/cm2 MaskedColumn 39420
line_hei_8446_err float64 MaskedColumn 39420
line_heii_4687 float64 Line flux of heii_4687 1e-20 erg/s/cm2 MaskedColumn 59306
line_heii_4687_err float64 MaskedColumn 59306
line_hg float64 Line flux of hg 1e-20 erg/s/cm2 MaskedColumn 38897
line_hg_err float64 MaskedColumn 38897
line_lya float64 Line flux of lya 1e-20 erg/s/cm2 MaskedColumn 68665
line_lya_err float64 MaskedColumn 68665
line_mgii float64 Line flux of mgii 1e-20 erg/s/cm2 MaskedColumn 49321
line_mgii_err float64 MaskedColumn 49321
line_neiii_3867 float64 Line flux of neiii_3867 1e-20 erg/s/cm2 MaskedColumn 61857
line_neiii_3867_err float64 MaskedColumn 61858
line_neiii_3968 float64 Line flux of neiii_3968 1e-20 erg/s/cm2 MaskedColumn 41226
line_neiii_3968_err float64 MaskedColumn 41228
line_nev_3346 float64 Line flux of nev_3346 1e-20 erg/s/cm2 MaskedColumn 45488
line_nev_3346_err float64 MaskedColumn 45488
line_nevi_3426 float64 Line flux of nevi_3426 1e-20 erg/s/cm2 MaskedColumn 44940
line_nevi_3426_err float64 MaskedColumn 44941
line_niii_1750 float64 Line flux of niii_1750 1e-20 erg/s/cm2 MaskedColumn 59941
line_niii_1750_err float64 MaskedColumn 59941
line_oi_6302 float64 Line flux of oi_6302 1e-20 erg/s/cm2 MaskedColumn 34327
line_oi_6302_err float64 MaskedColumn 34327
line_oii float64 Line flux of oii 1e-20 erg/s/cm2 MaskedColumn 42821
line_oii_7325 float64 Line flux of oii_7325 1e-20 erg/s/cm2 MaskedColumn 50553
line_oii_7325_err float64 MaskedColumn 50554
line_oii_err float64 MaskedColumn 42822
line_oiii float64 Line flux of combined OIII 4959+5007 1e-20 erg/s/cm2/A MaskedColumn 57718
line_oiii_1663 float64 Line flux of oiii_1663 1e-20 erg/s/cm2 MaskedColumn 61062
line_oiii_1663_err float64 MaskedColumn 61062
line_oiii_4363 float64 Line flux of oiii_4363 1e-20 erg/s/cm2 MaskedColumn 60229
line_oiii_4363_err float64 MaskedColumn 60229
line_oiii_4959 float64 Line flux of oiii_4959 1e-20 erg/s/cm2 MaskedColumn 58726
line_oiii_4959_err float64 MaskedColumn 58726
line_oiii_5007 float64 Line flux of oiii_5007 1e-20 erg/s/cm2 MaskedColumn 58598
line_oiii_5007_err float64 MaskedColumn 58598
line_oiii_err float64 MaskedColumn 57718
line_pa10 float64 Line flux of pa10 1e-20 erg/s/cm2 MaskedColumn 41147
line_pa10_err float64 MaskedColumn 41152
line_pa8 float64 Line flux of pa8 1e-20 erg/s/cm2 MaskedColumn 42686
line_pa8_err float64 MaskedColumn 42691
line_pa9 float64 Line flux of pa9 1e-20 erg/s/cm2 MaskedColumn 41764
line_pa9_err float64 MaskedColumn 41767
line_paa float64 Line flux of paa 1e-20 erg/s/cm2 MaskedColumn 63659
line_paa_err float64 MaskedColumn 63659
line_pab float64 Line flux of pab 1e-20 erg/s/cm2 MaskedColumn 52440
line_pab_err float64 MaskedColumn 52441
line_pad float64 Line flux of pad 1e-20 erg/s/cm2 MaskedColumn 44197
line_pad_err float64 MaskedColumn 44201
line_pag float64 Line flux of pag 1e-20 erg/s/cm2 MaskedColumn 46554
line_pag_err float64 MaskedColumn 46557
line_pfb float64 Line flux of pfb 1e-20 erg/s/cm2 MaskedColumn 79212
line_pfb_err float64 MaskedColumn 79212
line_pfd float64 Line flux of pfd 1e-20 erg/s/cm2 MaskedColumn 74917
line_pfd_err float64 MaskedColumn 74918
line_pfe float64 Line flux of pfe 1e-20 erg/s/cm2 MaskedColumn 73676
line_pfe_err float64 MaskedColumn 73676
line_pfg float64 Line flux of pfg 1e-20 erg/s/cm2 MaskedColumn 76553
line_pfg_err float64 MaskedColumn 76553
line_sii float64 Line flux of sii 1e-20 erg/s/cm2 MaskedColumn 49475
line_sii_err float64 MaskedColumn 49480
line_siii_9068 float64 Line flux of siii_9068 1e-20 erg/s/cm2 MaskedColumn 41324
line_siii_9068_err float64 MaskedColumn 41329
line_siii_9531 float64 Line flux of siii_9531 1e-20 erg/s/cm2 MaskedColumn 42649
line_siii_9531_err float64 MaskedColumn 42654
spl_0 float64 Spline continuum coefficient 0 MaskedColumn 18242
spl_0_err float64 MaskedColumn 18242
spl_1 float64 Spline continuum coefficient 1 MaskedColumn 18242
spl_10 float64 Spline continuum coefficient 10 MaskedColumn 18242
spl_10_err float64 MaskedColumn 18242
spl_11 float64 Spline continuum coefficient 11 MaskedColumn 18242
spl_11_err float64 MaskedColumn 18243
spl_12 float64 Spline continuum coefficient 12 MaskedColumn 18242
spl_12_err float64 MaskedColumn 18245
spl_13 float64 Spline continuum coefficient 13 MaskedColumn 18242
spl_13_err float64 MaskedColumn 18244
spl_14 float64 Spline continuum coefficient 14 MaskedColumn 18242
spl_14_err float64 MaskedColumn 18244
spl_15 float64 Spline continuum coefficient 15 MaskedColumn 18242
spl_15_err float64 MaskedColumn 18244
spl_16 float64 Spline continuum coefficient 16 MaskedColumn 18242
spl_16_err float64 MaskedColumn 18243
spl_17 float64 Spline continuum coefficient 17 MaskedColumn 18242
spl_17_err float64 MaskedColumn 18243
spl_18 float64 Spline continuum coefficient 18 MaskedColumn 18242
spl_18_err float64 MaskedColumn 18242
spl_19 float64 Spline continuum coefficient 19 MaskedColumn 18242
spl_19_err float64 MaskedColumn 18242
spl_1_err float64 MaskedColumn 18242
spl_2 float64 Spline continuum coefficient 2 MaskedColumn 18242
spl_20 float64 Spline continuum coefficient 20 MaskedColumn 18242
spl_20_err float64 MaskedColumn 18242
spl_21 float64 Spline continuum coefficient 21 MaskedColumn 18242
spl_21_err float64 MaskedColumn 18242
spl_22 float64 Spline continuum coefficient 22 MaskedColumn 18242
spl_22_err float64 MaskedColumn 18242
spl_2_err float64 MaskedColumn 18242
spl_3 float64 Spline continuum coefficient 3 MaskedColumn 18242
spl_3_err float64 MaskedColumn 18242
spl_4 float64 Spline continuum coefficient 4 MaskedColumn 18242
spl_4_err float64 MaskedColumn 18242
spl_5 float64 Spline continuum coefficient 5 MaskedColumn 18242
spl_5_err float64 MaskedColumn 18243
spl_6 float64 Spline continuum coefficient 6 MaskedColumn 18242
spl_6_err float64 MaskedColumn 18243
spl_7 float64 Spline continuum coefficient 7 MaskedColumn 18242
spl_7_err float64 MaskedColumn 18247
spl_8 float64 Spline continuum coefficient 8 MaskedColumn 18242
spl_8_err float64 MaskedColumn 18243
spl_9 float64 Spline continuum coefficient 9 MaskedColumn 18242
spl_9_err float64 MaskedColumn 18245
zline float64 Redshift where the lines where fit MaskedColumn 18242
line_civ_1549 float64 Line flux of civ_1549 1e-20 erg/s/cm2 MaskedColumn 62960
line_civ_1549_err float64 MaskedColumn 62960
line_h10 float64 Line flux of h10 1e-20 erg/s/cm2 MaskedColumn 71081
line_h10_err float64 MaskedColumn 71081
line_h11 float64 Line flux of h11 1e-20 erg/s/cm2 MaskedColumn 71209
line_h11_err float64 MaskedColumn 71209
line_h12 float64 Line flux of h12 1e-20 erg/s/cm2 MaskedColumn 71254
line_h12_err float64 MaskedColumn 71254
line_h7 float64 Line flux of h7 1e-20 erg/s/cm2 MaskedColumn 70536
line_h7_err float64 MaskedColumn 70536
line_h8 float64 Line flux of h8 1e-20 erg/s/cm2 MaskedColumn 70806
line_h8_err float64 MaskedColumn 70806
line_h9 float64 Line flux of h9 1e-20 erg/s/cm2 MaskedColumn 70958
line_h9_err float64 MaskedColumn 70958
line_ha float64 Line flux of ha 1e-20 erg/s/cm2 MaskedColumn 65641
line_ha_err float64 MaskedColumn 65641
line_hei_6680 float64 Line flux of hei_6680 1e-20 erg/s/cm2 MaskedColumn 65749
line_hei_6680_err float64 MaskedColumn 65749
line_heii_1640 float64 Line flux of heii_1640 1e-20 erg/s/cm2 MaskedColumn 61662
line_heii_1640_err float64 MaskedColumn 61662
line_nii_6549 float64 Line flux of nii_6549 1e-20 erg/s/cm2 MaskedColumn 65634
line_nii_6549_err float64 MaskedColumn 65634
line_nii_6584 float64 Line flux of nii_6584 1e-20 erg/s/cm2 MaskedColumn 65646
line_nii_6584_err float64 MaskedColumn 65646
line_oii_7323 float64 Line flux of oii_7323 1e-20 erg/s/cm2 MaskedColumn 66230
line_oii_7323_err float64 MaskedColumn 66230
line_oii_7332 float64 Line flux of oii_7332 1e-20 erg/s/cm2 MaskedColumn 66244
line_oii_7332_err float64 MaskedColumn 66244
line_sii_6717 float64 Line flux of sii_6717 1e-20 erg/s/cm2 MaskedColumn 65776
line_sii_6717_err float64 MaskedColumn 65776
line_sii_6731 float64 Line flux of sii_6731 1e-20 erg/s/cm2 MaskedColumn 65760
line_sii_6731_err float64 MaskedColumn 65760
line_siii_6314 float64 Line flux of siii_6314 1e-20 erg/s/cm2 MaskedColumn 65692
line_siii_6314_err float64 MaskedColumn 65692
escale0 float64 0th coefficient of the uncertainty scaling MaskedColumn 18242
escale1 float64 1st coefficient of the uncertainty scaling MaskedColumn 18242
line_ciii_1906 float64 Line flux of ciii_1906 1e-20 erg/s/cm2 MaskedColumn 57937
line_ciii_1906_err float64 MaskedColumn 57937
line_niv_1487 float64 Line flux of niv_1487 1e-20 erg/s/cm2 MaskedColumn 65762
line_niv_1487_err float64 MaskedColumn 65762
line_pah_3p29 float64 Line flux of pah_3p29 1e-20 erg/s/cm2 MaskedColumn 74940
line_pah_3p29_err float64 MaskedColumn 74940
line_pah_3p40 float64 Line flux of pah_3p40 1e-20 erg/s/cm2 MaskedColumn 74940
line_pah_3p40_err float64 MaskedColumn 74940
eqw_ariii_7138 float64 Observed-frame equivalent width in ariii_7138 MaskedColumn 35897
eqw_ariii_7753 float64 Observed-frame equivalent width in ariii_7753 MaskedColumn 37422
eqw_bra float64 Observed-frame equivalent width in bra MaskedColumn 77651
eqw_brb float64 Observed-frame equivalent width in brb MaskedColumn 71180
eqw_brd float64 Observed-frame equivalent width in brd MaskedColumn 64731
eqw_brg float64 Observed-frame equivalent width in brg MaskedColumn 67448
eqw_ciii_1906 float64 Observed-frame equivalent width in ciii_1906 MaskedColumn 57948
eqw_civ_1549 float64 Observed-frame equivalent width in civ_1549 MaskedColumn 62963
eqw_ha_nii float64 Observed-frame equivalent width in ha_nii MaskedColumn 49293
eqw_hb float64 Observed-frame equivalent width in hb MaskedColumn 36439
eqw_hd float64 Observed-frame equivalent width in hd MaskedColumn 40299
eqw_hei_1083 float64 Observed-frame equivalent width in hei_1083 MaskedColumn 46257
eqw_hei_3889 float64 Observed-frame equivalent width in hei_3889 MaskedColumn 51346
eqw_hei_5877 float64 Observed-frame equivalent width in hei_5877 MaskedColumn 34498
eqw_hei_7065 float64 Observed-frame equivalent width in hei_7065 MaskedColumn 35702
eqw_hei_8446 float64 Observed-frame equivalent width in hei_8446 MaskedColumn 39455
eqw_heii_1640 float64 Observed-frame equivalent width in heii_1640 MaskedColumn 61666
eqw_heii_4687 float64 Observed-frame equivalent width in heii_4687 MaskedColumn 59316
eqw_hg float64 Observed-frame equivalent width in hg MaskedColumn 38924
eqw_lya float64 Observed-frame equivalent width in lya MaskedColumn 68665
eqw_mgii float64 Observed-frame equivalent width in mgii MaskedColumn 49348
eqw_neiii_3867 float64 Observed-frame equivalent width in neiii_3867 MaskedColumn 61873
eqw_neiii_3968 float64 Observed-frame equivalent width in neiii_3968 MaskedColumn 41256
eqw_nev_3346 float64 Observed-frame equivalent width in nev_3346 MaskedColumn 45515
eqw_nevi_3426 float64 Observed-frame equivalent width in nevi_3426 MaskedColumn 44964
eqw_niii_1750 float64 Observed-frame equivalent width in niii_1750 MaskedColumn 59948
eqw_niv_1487 float64 Observed-frame equivalent width in niv_1487 MaskedColumn 65764
eqw_oi_6302 float64 Observed-frame equivalent width in oi_6302 MaskedColumn 34357
eqw_oii float64 Observed-frame equivalent width in oii MaskedColumn 42852
eqw_oii_7325 float64 Observed-frame equivalent width in oii_7325 MaskedColumn 50584
eqw_oiii float64 Observed-frame equivalent width in oiii MaskedColumn 57734
eqw_oiii_1663 float64 Observed-frame equivalent width in oiii_1663 MaskedColumn 61065
eqw_oiii_4363 float64 Observed-frame equivalent width in oiii_4363 MaskedColumn 60240
eqw_oiii_4959 float64 Observed-frame equivalent width in oiii_4959 MaskedColumn 58736
eqw_oiii_5007 float64 Observed-frame equivalent width in oiii_5007 MaskedColumn 58608
eqw_pa10 float64 Observed-frame equivalent width in pa10 MaskedColumn 41179
eqw_pa8 float64 Observed-frame equivalent width in pa8 MaskedColumn 42725
eqw_pa9 float64 Observed-frame equivalent width in pa9 MaskedColumn 41798
eqw_paa float64 Observed-frame equivalent width in paa MaskedColumn 63671
eqw_pab float64 Observed-frame equivalent width in pab MaskedColumn 52477
eqw_pad float64 Observed-frame equivalent width in pad MaskedColumn 44239
eqw_pag float64 Observed-frame equivalent width in pag MaskedColumn 46592
eqw_pfb float64 Observed-frame equivalent width in pfb MaskedColumn 79212
eqw_pfd float64 Observed-frame equivalent width in pfd MaskedColumn 74920
eqw_pfe float64 Observed-frame equivalent width in pfe MaskedColumn 73680
eqw_pfg float64 Observed-frame equivalent width in pfg MaskedColumn 76556
eqw_sii float64 Observed-frame equivalent width in sii MaskedColumn 49501
eqw_siii_9068 float64 Observed-frame equivalent width in siii_9068 MaskedColumn 41361
eqw_siii_9531 float64 Observed-frame equivalent width in siii_9531 MaskedColumn 42685
line_ha_nii float64 Line flux of combined Ha+NII with 3:1 ratio MaskedColumn 49269
line_ha_nii_err float64 MaskedColumn 49273
eqw_h10 float64 Observed-frame equivalent width in h10 MaskedColumn 71081
eqw_h11 float64 Observed-frame equivalent width in h11 MaskedColumn 71209
eqw_h12 float64 Observed-frame equivalent width in h12 MaskedColumn 71254
eqw_h7 float64 Observed-frame equivalent width in h7 MaskedColumn 70536
eqw_h8 float64 Observed-frame equivalent width in h8 MaskedColumn 70806
eqw_h9 float64 Observed-frame equivalent width in h9 MaskedColumn 70958
eqw_ha float64 Observed-frame equivalent width in ha MaskedColumn 65643
eqw_hei_6680 float64 Observed-frame equivalent width in hei_6680 MaskedColumn 65749
eqw_nii_6549 float64 Observed-frame equivalent width in nii_6549 MaskedColumn 65635
eqw_nii_6584 float64 Observed-frame equivalent width in nii_6584 MaskedColumn 65647
eqw_oii_7323 float64 Observed-frame equivalent width in oii_7323 MaskedColumn 66230
eqw_oii_7332 float64 Observed-frame equivalent width in oii_7332 MaskedColumn 66244
eqw_sii_6717 float64 Observed-frame equivalent width in sii_6717 MaskedColumn 65776
eqw_sii_6731 float64 Observed-frame equivalent width in sii_6731 MaskedColumn 65760
eqw_siii_6314 float64 Observed-frame equivalent width in siii_6314 MaskedColumn 65692
sn_line float64 .1f Maximum emission line SN MaskedColumn 18242
ztime float64 UNIX time of redshift fit MaskedColumn 18242
line_ci_9850 float64 Line flux of ci_9850 1e-20 erg/s/cm2 MaskedColumn 43554
line_ci_9850_err float64 MaskedColumn 43558
line_feii_11128 float64 Line flux of feii_11128 1e-20 erg/s/cm2 MaskedColumn 47164
line_feii_11128_err float64 MaskedColumn 47167
line_pii_11886 float64 Line flux of pii_11886 1e-20 erg/s/cm2 MaskedColumn 49532
line_pii_11886_err float64 MaskedColumn 49532
line_feii_12570 float64 Line flux of feii_12570 1e-20 erg/s/cm2 MaskedColumn 51790
line_feii_12570_err float64 MaskedColumn 51790
eqw_ci_9850 float64 Observed-frame equivalent width in ci_9850 MaskedColumn 43588
eqw_feii_11128 float64 Observed-frame equivalent width in feii_11128 MaskedColumn 47206
eqw_pii_11886 float64 Observed-frame equivalent width in pii_11886 MaskedColumn 49563
eqw_feii_12570 float64 Observed-frame equivalent width in feii_12570 MaskedColumn 51827
line_feii_16440 float64 Line flux of feii_16440 1e-20 erg/s/cm2 MaskedColumn 59928
line_feii_16440_err float64 MaskedColumn 59928
line_feii_16877 float64 Line flux of feii_16877 1e-20 erg/s/cm2 MaskedColumn 60978
line_feii_16877_err float64 MaskedColumn 60978
line_brf float64 Line flux of brf 1e-20 erg/s/cm2 MaskedColumn 61730
line_brf_err float64 MaskedColumn 61730
line_feii_17418 float64 Line flux of feii_17418 1e-20 erg/s/cm2 MaskedColumn 61813
line_feii_17418_err float64 MaskedColumn 61813
line_bre float64 Line flux of bre 1e-20 erg/s/cm2 MaskedColumn 62881
line_bre_err float64 MaskedColumn 62882
line_feii_18362 float64 Line flux of feii_18362 1e-20 erg/s/cm2 MaskedColumn 63211
line_feii_18362_err float64 MaskedColumn 63211
eqw_feii_16440 float64 Observed-frame equivalent width in feii_16440 MaskedColumn 59946
eqw_feii_16877 float64 Observed-frame equivalent width in feii_16877 MaskedColumn 60995
eqw_brf float64 Observed-frame equivalent width in brf MaskedColumn 61749
eqw_feii_17418 float64 Observed-frame equivalent width in feii_17418 MaskedColumn 61829
eqw_bre float64 Observed-frame equivalent width in bre MaskedColumn 62895
eqw_feii_18362 float64 Observed-frame equivalent width in feii_18362 MaskedColumn 63227
valid str5 Redshift matches best z from visual inspection MaskedColumn 38983
objid int64 Unique source identifier Column 0
z_best float64 Best redshift estimate for unique sources Column 0
ztype str1 Source for z_best (G)rating or (P)rism MaskedColumn 38728
z_prism float64 Best redshift estimate from prism spectra Column 0
z_grating float64 Best redshift estiamte from grating spectra Column 0
phot_correction float64 .2f Scale to photometry -log10(c) = -0.902 log10(flux_radius) + 0.649 log10(profsig) + 0.605 MaskedColumn 29922
phot_flux_radius float64 .2f FLUX_RADIUS from photometric source MaskedColumn 29922
phot_dr float64 Offset to the nearest source in the photometric catalog in arcsec MaskedColumn 29922
file_phot str44 Filename of the photometric catalog MaskedColumn 29922
id_phot int64 ID number in the photometric cadtalog MaskedColumn 29922
phot_mag_auto float64 .2f Kron MAG_AUTO in the photometric detection image MaskedColumn 29922
phot_f090w_tot_1 float64 Total f090w flux density from the photometric catalog MaskedColumn 29922
phot_f090w_etot_1 float64 MaskedColumn 29922
phot_f115w_tot_1 float64 Total f115w flux density from the photometric catalog MaskedColumn 29922
phot_f115w_etot_1 float64 MaskedColumn 29922
phot_f150w_tot_1 float64 Total f150w flux density from the photometric catalog MaskedColumn 29922
phot_f150w_etot_1 float64 MaskedColumn 29922
phot_f200w_tot_1 float64 Total f200w flux density from the photometric catalog MaskedColumn 29922
phot_f200w_etot_1 float64 MaskedColumn 29922
phot_f277w_tot_1 float64 Total f277w flux density from the photometric catalog MaskedColumn 29922
phot_f277w_etot_1 float64 MaskedColumn 29922
phot_f356w_tot_1 float64 Total f356w flux density from the photometric catalog MaskedColumn 29922
phot_f356w_etot_1 float64 MaskedColumn 29922
phot_f410m_tot_1 float64 Total f410m flux density from the photometric catalog MaskedColumn 29922
phot_f410m_etot_1 float64 MaskedColumn 29922
phot_f444w_tot_1 float64 Total f444w flux density from the photometric catalog MaskedColumn 29922
phot_f444w_etot_1 float64 MaskedColumn 29922
phot_Av float64 Av from the photoz fit MaskedColumn 29922
phot_mass float64 stellar mass from the photoz fit MaskedColumn 29922
phot_restU float64 flux density of the redshifted U band from the photoz fit MaskedColumn 29922
phot_restV float64 flux density of the redshifted V band from the photoz fit MaskedColumn 29922
phot_restJ float64 flux density of the redshifted J band from the photoz fit MaskedColumn 29922
z_phot float64 photometric redshift MaskedColumn 29922
phot_LHa float64 Halpha luminosity from the photo-z fit MaskedColumn 29922
phot_LOIII float64 OIII luminosity from the photo-z fit MaskedColumn 29922
phot_LOII float64 OII luminosity from the photo-z fit MaskedColumn 29922
grade int64 Grade from visual inspection MaskedColumn 38015
zgrade float64 9.5f Redshift from visual inspection MaskedColumn 38015
reviewer str4 Initials of the visual inspection reviewer MaskedColumn 38015
comment str93 Comment from visual inspection MaskedColumn 48371
zrf float64 Redshift used for integrated rest-frame filter MaskedColumn 18242
escale float64 MaskedColumn 18242
obs_239_valid int64 MaskedColumn 18242
obs_239_frac float64 Fraction of wfc_f814w_t81.dat covered by spectrum MaskedColumn 18242
obs_239_flux float64 Spectrum flux in wfc_f814w_t81.dat MaskedColumn 18242
obs_239_err float64 Spectrum err in wfc_f814w_t81.dat MaskedColumn 18242
obs_239_full_err float64 Spectrum err in wfc_f814w_t81.dat MaskedColumn 18242
obs_205_valid int64 MaskedColumn 18242
obs_205_frac float64 Fraction of f160w.dat covered by spectrum MaskedColumn 18242
obs_205_flux float64 Spectrum flux in f160w.dat MaskedColumn 18242
obs_205_err float64 Spectrum err in f160w.dat MaskedColumn 18242
obs_205_full_err float64 Spectrum err in f160w.dat MaskedColumn 18242
obs_362_valid int64 MaskedColumn 18242
obs_362_frac float64 Fraction of jwst_nircam_f070w covered by spectrum MaskedColumn 18242
obs_362_flux float64 Spectrum flux in jwst_nircam_f070w MaskedColumn 18242
obs_362_err float64 Spectrum err in jwst_nircam_f070w MaskedColumn 18242
obs_362_full_err float64 Spectrum err in jwst_nircam_f070w MaskedColumn 18242
obs_363_valid int64 MaskedColumn 18242
obs_363_frac float64 Fraction of jwst_nircam_f090w covered by spectrum MaskedColumn 18242
obs_363_flux float64 Spectrum flux in jwst_nircam_f090w MaskedColumn 18242
obs_363_err float64 Spectrum err in jwst_nircam_f090w MaskedColumn 18242
obs_363_full_err float64 Spectrum err in jwst_nircam_f090w MaskedColumn 18242
obs_364_valid int64 MaskedColumn 18242
obs_364_frac float64 Fraction of jwst_nircam_f115w covered by spectrum MaskedColumn 18242
obs_364_flux float64 Spectrum flux in jwst_nircam_f115w MaskedColumn 18242
obs_364_err float64 Spectrum err in jwst_nircam_f115w MaskedColumn 18242
obs_364_full_err float64 Spectrum err in jwst_nircam_f115w MaskedColumn 18242
obs_365_valid int64 MaskedColumn 18242
obs_365_frac float64 Fraction of jwst_nircam_f150w covered by spectrum MaskedColumn 18242
obs_365_flux float64 Spectrum flux in jwst_nircam_f150w MaskedColumn 18242
obs_365_err float64 Spectrum err in jwst_nircam_f150w MaskedColumn 18242
obs_365_full_err float64 Spectrum err in jwst_nircam_f150w MaskedColumn 18242
obs_366_valid int64 MaskedColumn 18242
obs_366_frac float64 Fraction of jwst_nircam_f200w covered by spectrum MaskedColumn 18242
obs_366_flux float64 Spectrum flux in jwst_nircam_f200w MaskedColumn 18242
obs_366_err float64 Spectrum err in jwst_nircam_f200w MaskedColumn 18242
obs_366_full_err float64 Spectrum err in jwst_nircam_f200w MaskedColumn 18242
obs_370_valid int64 MaskedColumn 18242
obs_370_frac float64 Fraction of jwst_nircam_f182m covered by spectrum MaskedColumn 18242
obs_370_flux float64 Spectrum flux in jwst_nircam_f182m MaskedColumn 18242
obs_370_err float64 Spectrum err in jwst_nircam_f182m MaskedColumn 18242
obs_370_full_err float64 Spectrum err in jwst_nircam_f182m MaskedColumn 18242
obs_371_valid int64 MaskedColumn 18242
obs_371_frac float64 Fraction of jwst_nircam_f210m covered by spectrum MaskedColumn 18242
obs_371_flux float64 Spectrum flux in jwst_nircam_f210m MaskedColumn 18242
obs_371_err float64 Spectrum err in jwst_nircam_f210m MaskedColumn 18242
obs_371_full_err float64 Spectrum err in jwst_nircam_f210m MaskedColumn 18242
obs_375_valid int64 MaskedColumn 18242
obs_375_frac float64 Fraction of jwst_nircam_f277w covered by spectrum MaskedColumn 18242
obs_375_flux float64 Spectrum flux in jwst_nircam_f277w MaskedColumn 18242
obs_375_err float64 Spectrum err in jwst_nircam_f277w MaskedColumn 18242
obs_375_full_err float64 Spectrum err in jwst_nircam_f277w MaskedColumn 18242
obs_376_valid int64 MaskedColumn 18242
obs_376_frac float64 Fraction of jwst_nircam_f356w covered by spectrum MaskedColumn 18242
obs_376_flux float64 Spectrum flux in jwst_nircam_f356w MaskedColumn 18242
obs_376_err float64 Spectrum err in jwst_nircam_f356w MaskedColumn 18242
obs_376_full_err float64 Spectrum err in jwst_nircam_f356w MaskedColumn 18242
obs_377_valid int64 MaskedColumn 18242
obs_377_frac float64 Fraction of jwst_nircam_f444w covered by spectrum MaskedColumn 18242
obs_377_flux float64 Spectrum flux in jwst_nircam_f444w MaskedColumn 18242
obs_377_err float64 Spectrum err in jwst_nircam_f444w MaskedColumn 18242
obs_377_full_err float64 Spectrum err in jwst_nircam_f444w MaskedColumn 18242
obs_379_valid int64 MaskedColumn 18242
obs_379_frac float64 Fraction of jwst_nircam_f250m covered by spectrum MaskedColumn 18242
obs_379_flux float64 Spectrum flux in jwst_nircam_f250m MaskedColumn 18242
obs_379_err float64 Spectrum err in jwst_nircam_f250m MaskedColumn 18242
obs_379_full_err float64 Spectrum err in jwst_nircam_f250m MaskedColumn 18242
obs_380_valid int64 MaskedColumn 18242
obs_380_frac float64 Fraction of jwst_nircam_f300m covered by spectrum MaskedColumn 18242
obs_380_flux float64 Spectrum flux in jwst_nircam_f300m MaskedColumn 18242
obs_380_err float64 Spectrum err in jwst_nircam_f300m MaskedColumn 18242
obs_380_full_err float64 Spectrum err in jwst_nircam_f300m MaskedColumn 18242
obs_381_valid int64 MaskedColumn 18242
obs_381_frac float64 Fraction of jwst_nircam_f335m covered by spectrum MaskedColumn 18242
obs_381_flux float64 Spectrum flux in jwst_nircam_f335m MaskedColumn 18242
obs_381_err float64 Spectrum err in jwst_nircam_f335m MaskedColumn 18242
obs_381_full_err float64 Spectrum err in jwst_nircam_f335m MaskedColumn 18242
obs_382_valid int64 MaskedColumn 18242
obs_382_frac float64 Fraction of jwst_nircam_f360m covered by spectrum MaskedColumn 18242
obs_382_flux float64 Spectrum flux in jwst_nircam_f360m MaskedColumn 18242
obs_382_err float64 Spectrum err in jwst_nircam_f360m MaskedColumn 18242
obs_382_full_err float64 Spectrum err in jwst_nircam_f360m MaskedColumn 18242
obs_383_valid int64 MaskedColumn 18242
obs_383_frac float64 Fraction of jwst_nircam_f410m covered by spectrum MaskedColumn 18242
obs_383_flux float64 Spectrum flux in jwst_nircam_f410m MaskedColumn 18242
obs_383_err float64 Spectrum err in jwst_nircam_f410m MaskedColumn 18242
obs_383_full_err float64 Spectrum err in jwst_nircam_f410m MaskedColumn 18242
obs_384_valid int64 MaskedColumn 18242
obs_384_frac float64 Fraction of jwst_nircam_f430m covered by spectrum MaskedColumn 18242
obs_384_flux float64 Spectrum flux in jwst_nircam_f430m MaskedColumn 18242
obs_384_err float64 Spectrum err in jwst_nircam_f430m MaskedColumn 18242
obs_384_full_err float64 Spectrum err in jwst_nircam_f430m MaskedColumn 18242
obs_385_valid int64 MaskedColumn 18242
obs_385_frac float64 Fraction of jwst_nircam_f460m covered by spectrum MaskedColumn 18242
obs_385_flux float64 Spectrum flux in jwst_nircam_f460m MaskedColumn 18242
obs_385_err float64 Spectrum err in jwst_nircam_f460m MaskedColumn 18242
obs_385_full_err float64 Spectrum err in jwst_nircam_f460m MaskedColumn 18242
obs_386_valid int64 MaskedColumn 18242
obs_386_frac float64 Fraction of jwst_nircam_f480m covered by spectrum MaskedColumn 18242
obs_386_flux float64 Spectrum flux in jwst_nircam_f480m MaskedColumn 18242
obs_386_err float64 Spectrum err in jwst_nircam_f480m MaskedColumn 18242
obs_386_full_err float64 Spectrum err in jwst_nircam_f480m MaskedColumn 18242
rest_120_valid int64 MaskedColumn 18242
rest_120_frac float64 Fraction of galex1500.res covered by spectrum MaskedColumn 18242
rest_120_flux float64 Spectrum flux in galex1500.res MaskedColumn 18242
rest_120_err float64 Spectrum err in galex1500.res MaskedColumn 18242
rest_120_full_err float64 Spectrum err in galex1500.res MaskedColumn 18242
rest_121_valid int64 MaskedColumn 18242
rest_121_frac float64 Fraction of galex2500.res covered by spectrum MaskedColumn 18242
rest_121_flux float64 Spectrum flux in galex2500.res MaskedColumn 18242
rest_121_err float64 Spectrum err in galex2500.res MaskedColumn 18242
rest_121_full_err float64 Spectrum err in galex2500.res MaskedColumn 18242
rest_218_valid int64 MaskedColumn 18242
rest_218_frac float64 Fraction of UV1600.dat covered by spectrum MaskedColumn 18242
rest_218_flux float64 Spectrum flux in UV1600.dat MaskedColumn 18242
rest_218_err float64 Spectrum err in UV1600.dat MaskedColumn 18242
rest_218_full_err float64 Spectrum err in UV1600.dat MaskedColumn 18242
rest_219_valid int64 MaskedColumn 18242
rest_219_frac float64 Fraction of UV2800.dat covered by spectrum MaskedColumn 18242
rest_219_flux float64 Spectrum flux in UV2800.dat MaskedColumn 18242
rest_219_err float64 Spectrum err in UV2800.dat MaskedColumn 18242
rest_219_full_err float64 Spectrum err in UV2800.dat MaskedColumn 18242
rest_270_valid int64 MaskedColumn 18242
rest_270_frac float64 Fraction of Tophat_1400_200.dat covered by spectrum MaskedColumn 18242
rest_270_flux float64 Spectrum flux in Tophat_1400_200.dat MaskedColumn 18242
rest_270_err float64 Spectrum err in Tophat_1400_200.dat MaskedColumn 18242
rest_270_full_err float64 Spectrum err in Tophat_1400_200.dat MaskedColumn 18242
rest_271_valid int64 MaskedColumn 18242
rest_271_frac float64 Fraction of Tophat_1700_200.dat covered by spectrum MaskedColumn 18242
rest_271_flux float64 Spectrum flux in Tophat_1700_200.dat MaskedColumn 18242
rest_271_err float64 Spectrum err in Tophat_1700_200.dat MaskedColumn 18242
rest_271_full_err float64 Spectrum err in Tophat_1700_200.dat MaskedColumn 18242
rest_272_valid int64 MaskedColumn 18242
rest_272_frac float64 Fraction of Tophat_2200_200.dat covered by spectrum MaskedColumn 18242
rest_272_flux float64 Spectrum flux in Tophat_2200_200.dat MaskedColumn 18242
rest_272_err float64 Spectrum err in Tophat_2200_200.dat MaskedColumn 18242
rest_272_full_err float64 Spectrum err in Tophat_2200_200.dat MaskedColumn 18242
rest_274_valid int64 MaskedColumn 18242
rest_274_frac float64 Fraction of Tophat_2800_200.dat covered by spectrum MaskedColumn 18242
rest_274_flux float64 Spectrum flux in Tophat_2800_200.dat MaskedColumn 18242
rest_274_err float64 Spectrum err in Tophat_2800_200.dat MaskedColumn 18242
rest_274_full_err float64 Spectrum err in Tophat_2800_200.dat MaskedColumn 18242
rest_153_valid int64 MaskedColumn 18242
rest_153_frac float64 Fraction of maiz-apellaniz_Johnson_U.res covered by spectrum MaskedColumn 18242
rest_153_flux float64 Spectrum flux in maiz-apellaniz_Johnson_U.res MaskedColumn 18242
rest_153_err float64 Spectrum err in maiz-apellaniz_Johnson_U.res MaskedColumn 18242
rest_153_full_err float64 Spectrum err in maiz-apellaniz_Johnson_U.res MaskedColumn 18242
rest_154_valid int64 MaskedColumn 18242
rest_154_frac float64 Fraction of maiz-apellaniz_Johnson_B.res covered by spectrum MaskedColumn 18242
rest_154_flux float64 Spectrum flux in maiz-apellaniz_Johnson_B.res MaskedColumn 18242
rest_154_err float64 Spectrum err in maiz-apellaniz_Johnson_B.res MaskedColumn 18242
rest_154_full_err float64 Spectrum err in maiz-apellaniz_Johnson_B.res MaskedColumn 18242
rest_155_valid int64 MaskedColumn 18242
rest_155_frac float64 Fraction of maiz-apellaniz_Johnson_V.res covered by spectrum MaskedColumn 18242
rest_155_flux float64 Spectrum flux in maiz-apellaniz_Johnson_V.res MaskedColumn 18242
rest_155_err float64 Spectrum err in maiz-apellaniz_Johnson_V.res MaskedColumn 18242
rest_155_full_err float64 Spectrum err in maiz-apellaniz_Johnson_V.res MaskedColumn 18242
rest_156_valid int64 MaskedColumn 18242
rest_156_frac float64 Fraction of u.dat covered by spectrum MaskedColumn 18242
rest_156_flux float64 Spectrum flux in u.dat MaskedColumn 18242
rest_156_err float64 Spectrum err in u.dat MaskedColumn 18242
rest_156_full_err float64 Spectrum err in u.dat MaskedColumn 18242
rest_157_valid int64 MaskedColumn 18242
rest_157_frac float64 Fraction of g.dat covered by spectrum MaskedColumn 18242
rest_157_flux float64 Spectrum flux in g.dat MaskedColumn 18242
rest_157_err float64 Spectrum err in g.dat MaskedColumn 18242
rest_157_full_err float64 Spectrum err in g.dat MaskedColumn 18242
rest_158_valid int64 MaskedColumn 18242
rest_158_frac float64 Fraction of r.dat covered by spectrum MaskedColumn 18242
rest_158_flux float64 Spectrum flux in r.dat MaskedColumn 18242
rest_158_err float64 Spectrum err in r.dat MaskedColumn 18242
rest_158_full_err float64 Spectrum err in r.dat MaskedColumn 18242
rest_159_valid int64 MaskedColumn 18242
rest_159_frac float64 Fraction of i.dat covered by spectrum MaskedColumn 18242
rest_159_flux float64 Spectrum flux in i.dat MaskedColumn 18242
rest_159_err float64 Spectrum err in i.dat MaskedColumn 18242
rest_159_full_err float64 Spectrum err in i.dat MaskedColumn 18242
rest_160_valid int64 MaskedColumn 18242
rest_160_frac float64 Fraction of z.dat covered by spectrum MaskedColumn 18242
rest_160_flux float64 Spectrum flux in z.dat MaskedColumn 18242
rest_160_err float64 Spectrum err in z.dat MaskedColumn 18242
rest_160_full_err float64 Spectrum err in z.dat MaskedColumn 18242
rest_161_valid int64 MaskedColumn 18242
rest_161_frac float64 Fraction of J.res covered by spectrum MaskedColumn 18242
rest_161_flux float64 Spectrum flux in J.res MaskedColumn 18242
rest_161_err float64 Spectrum err in J.res MaskedColumn 18242
rest_161_full_err float64 Spectrum err in J.res MaskedColumn 18242
rest_162_valid int64 MaskedColumn 18242
rest_162_frac float64 Fraction of H.res covered by spectrum MaskedColumn 18242
rest_162_flux float64 Spectrum flux in H.res MaskedColumn 18242
rest_162_err float64 Spectrum err in H.res MaskedColumn 18242
rest_162_full_err float64 Spectrum err in H.res MaskedColumn 18242
rest_163_valid int64 MaskedColumn 18242
rest_163_frac float64 Fraction of K.res covered by spectrum MaskedColumn 18242
rest_163_flux float64 Spectrum flux in K.res MaskedColumn 18242
rest_163_err float64 Spectrum err in K.res MaskedColumn 18242
rest_163_full_err float64 Spectrum err in K.res MaskedColumn 18242
rest_414_valid int64 MaskedColumn 18242
rest_414_frac float64 Fraction of synthetic_u covered by spectrum MaskedColumn 18242
rest_414_flux float64 Spectrum flux in synthetic_u MaskedColumn 18242
rest_414_err float64 Spectrum err in synthetic_u MaskedColumn 18242
rest_414_full_err float64 Spectrum err in synthetic_u MaskedColumn 18242
rest_415_valid int64 MaskedColumn 18242
rest_415_frac float64 Fraction of synthetic_g covered by spectrum MaskedColumn 18242
rest_415_flux float64 Spectrum flux in synthetic_g MaskedColumn 18242
rest_415_err float64 Spectrum err in synthetic_g MaskedColumn 18242
rest_415_full_err float64 Spectrum err in synthetic_g MaskedColumn 18242
rest_416_valid int64 MaskedColumn 18242
rest_416_frac float64 Fraction of synthetic_i covered by spectrum MaskedColumn 18242
rest_416_flux float64 Spectrum flux in synthetic_i MaskedColumn 18242
rest_416_err float64 Spectrum err in synthetic_i MaskedColumn 18242
rest_416_full_err float64 Spectrum err in synthetic_i MaskedColumn 18242
beta float64 Estimated UV slope beta MaskedColumn 51222
beta_ref_flux float64 MaskedColumn 51221
beta_npix int64 Number of pixels for beta fit MaskedColumn 18242
beta_wlo float64 Minimum wavelength used for beta fit MaskedColumn 51221
beta_whi float64 Maximum wavelength used for beta fit MaskedColumn 51221
beta_nmad float64 NMAD of the beta fit MaskedColumn 51225
dla_npix float64 Number of pixels for the DLA fit MaskedColumn 51221
dla_value float64 DLA parameter from Heintz et al. MaskedColumn 51221
dla_unc float64 Uncertainty on DLA parameter MaskedColumn 51221
beta_cov_00 float64 Components of the beta covariance matrix MaskedColumn 51221
beta_cov_01 float64 Components of the beta covariance matrix MaskedColumn 51221
beta_cov_10 float64 Components of the beta covariance matrix MaskedColumn 51221
beta_cov_11 float64 Components of the beta covariance matrix MaskedColumn 51221
Add some preview columns to the table
RGB_URL = "https://grizli-cutout.herokuapp.com/thumb?size=1.5&scl=2.0&asinh=True&filters=f115w-clear%2Cf277w-clear%2Cf444w-clear&rgb_scl=1.5%2C0.74%2C1.3&pl=2&coord={ra}%2C{dec}"
tab['metafile'] = [m.split('_')[0] for m in tab['msamet']]
SLIT_URL = "https://grizli-cutout.herokuapp.com/thumb?size=1.5&scl=4.0&invert=True&filters=f444w-clear&rgb_scl=1.5%2C0.74%2C1.3&pl=2&coord={ra}%2C{dec}&nirspec=True&dpi_scale=6&nrs_lw=0.5&nrs_alpha=0.8&metafile={metafile}"
FITS_URL = "https://s3.amazonaws.com/msaexp-nirspec/extractions/{root}/{file}"
tab['Thumb'] = [
"<img src=\"{0}\" height=200px>".format(
RGB_URL.format(**row['ra','dec'])
)
for row in tab
]
tab['Slit_Thumb'] = [
"<img src=\"{0}\" height=200px>".format(
SLIT_URL.format(**row['ra','dec','metafile'])
)
for row in tab
]
tab['Spectrum_fnu'] = [
"<img src=\"{0}\" height=200px>".format(
FITS_URL.format(**row['root','file']).replace('.spec.fits', '.fnu.png')
)
for row in tab
]
tab['Spectrum_flam'] = [
"<img src=\"{0}\" height=200px>".format(
FITS_URL.format(**row['root','file']).replace('.spec.fits', '.flam.png')
)
for row in tab
]
# CANUCS in a different bucket
canucs = np.isin(
tab['root'],
['abell370-v4', 'macs0416-v4', 'macs0417-v4', 'macs1149-v4', 'macs1423-v4']
)
for j in tqdm(np.where(canucs)[0]):
for c in ['Spectrum_fnu', 'Spectrum_flam']:
tab[c][j] = tab[c][j].replace(
'msaexp-nirspec/extractions',
'grizli-canucs/nirspec'
)
100%|██████████████████████████████████████████████████████████████████████████████████████████████████████████| 1534/1534 [00:00<00:00, 97100.33it/s]
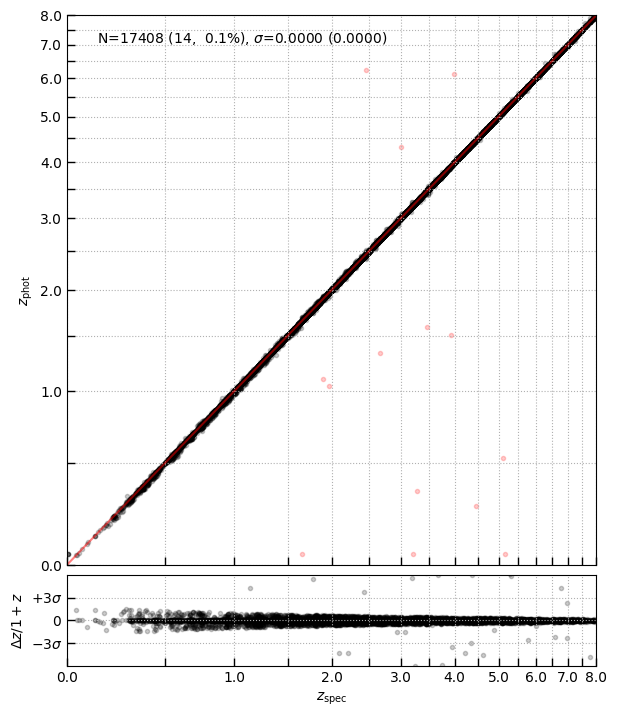
zphot - zspec
Compare the “best” NIRSpec redshift with grade=3 (grating if available, prism otherwise) to the photometric redshift in the matched catalogs.
import eazy.utils
test = (tab['grade'] == 3) & (tab['z_phot'].filled(-1.) > 0)
test &= (tab['grating'] == 'PRISM')
print(test.sum())
_ = eazy.utils.zphot_zspec(tab['z_phot'][test], tab['z_best'][test], zmax=14)
13703
# Counts by mask / program
utils.Unique(tab['root'], sort_counts=False)
N value
==== ==========
314 abell2744-castellano1-v4
613 abell2744-castellano2-v4
114 abell2744-ddt-v4
462 abell2744-glass-v4
299 abell370-v4
141 aurora-gdn01-v4
153 aurora-gdn02-v4
9 bd-ic348-gto-v4
25 bd-orion-gto-v4
344 bd-orion-gto2-v4
219 bluejay-north-v4
224 bluejay-south-v4
52 borg-0037m3337-v4
39 borg-0314m6712-v4
37 borg-0409m5317-v4
36 borg-0440m5244-v4
38 borg-0859p4114-v4
48 borg-0955p4528-v4
38 borg-1033p5051-v4
43 borg-1437p5044-v4
31 borg-2203p1851-v4
1107 cal-m31-pn2538-v4
414 cal-ocen-degraaff-v4
63 cantalupo-filament-02-v4
338 capers-cos01-v4
286 capers-cos04-v4
377 capers-cos07-v4
417 capers-cos10-v4
292 capers-cos13-v4
306 capers-cos16-v4
377 capers-cos19-v4
376 capers-egs44-v4
396 capers-egs47-v4
408 capers-egs49-v4
415 capers-egs53-v4
328 capers-egs55-v4
389 capers-egs61-v4
372 capers-egs65-v4
351 capers-udsp1-v4
302 capers-udsp2-v4
281 capers-udsp3-v4
170 capers-udsp5-v4
102 cecilia-v4
259 ceers-ddt-v4
1818 ceers-v4
96 cosmos-alpha-v4
367 cosmos-curti-v4
71 cosmos-lae-martin-v4
307 cosmos-transients-v4
84 cristal-cos01-v4
342 egs-mason-v4
272 egs-nelsonx-v4
127 excels-uds01-v4
128 excels-uds02-v4
136 excels-uds03-v4
142 excels-uds04-v4
44 gdn-chisholm-v4
580 gdn-fujimoto-v4
190 gdn-pah123-v4
66 gdn-pah4-v4
57 gds-barrufet-s156-v4
82 gds-barrufet-s67-v4
1236 gds-deep-v4
356 gds-egami-ddt-v4
256 gds-maseda-v4
724 gds-rieke-v4
825 gds-udeep-v4
96 glazebrook-cos-obs1-v4
110 glazebrook-cos-obs2-v4
108 glazebrook-cos-obs3-v4
122 glazebrook-egs-v4
238 glazebrook-v4
129 glimpse-obs01-v4
118 glimpse-obs01b-v4
135 glimpse-obs02-v4
192 goodsn-wide-v4
594 goodsn-wide0-v4
198 goodsn-wide1-v4
581 goodsn-wide2-v4
587 goodsn-wide3-v4
565 goodsn-wide6-v4
581 goodsn-wide66-v4
567 goodsn-wide7-v4
572 goodsn-wide8-v4
610 gto-wide-cos01-v4
570 gto-wide-cos02-v4
589 gto-wide-cos03-v4
575 gto-wide-cos04-v4
573 gto-wide-cos05-v4
2034 gto-wide-egs1-v4
594 gto-wide-egs2-v4
596 gto-wide-uds10-v4
592 gto-wide-uds11-v4
594 gto-wide-uds12-v4
609 gto-wide-uds13-v4
583 gto-wide-uds14-v4
126 iras16293-v4
512 j0226-wang-v4
161 j0252m0503-hennawi-02-v4
159 j0252m0503-hennawi-07-v4
399 j0910-wang-v4
234 j1007p2115-hennawi-v4
92 j1148-eilers-v4
34 j1342-msa-v4
774 jades-gdn-v4
1253 jades-gdn09-v4
1286 jades-gdn10-v4
1284 jades-gdn11-v4
837 jades-gdn198-v4
3473 jades-gdn2-v4
570 jades-gds-w03-v4
600 jades-gds-w04-v4
578 jades-gds-w05-v4
579 jades-gds-w06-v4
570 jades-gds-w07-v4
588 jades-gds-w08-v4
724 jades-gds-w09-v4
2979 jades-gds-wide-v4
1853 jades-gds-wide2-v4
953 jades-gds-wide3-v4
983 jades-gds02-v4
970 jades-gds03-v4
1014 jades-gds04-v4
1148 jades-gds05-v4
968 jades-gds06-v4
978 jades-gds07-v4
1008 jades-gds08-v4
924 jades-gds1-v4
835 jades-gds10-v4
100 lyc22-schaerer-01-v4
80 lyc22-schaerer-03-v4
104 lyc22-schaerer-12-v4
583 macs0416-nakajima-v4
284 macs0416-v4
318 macs0417-v4
404 macs1149-stiavelli-v4
149 macs1149-stiavelli2-v4
381 macs1149-v4
252 macs1423-v4
44 macsj0647-hr-v4
137 macsj0647-single-v4
142 macsj0647-v4
178 mom-uds01-v4
182 mom-uds02-v4
755 nexus-obs3-v4
958 nexus-obs5-v4
426 ngc628-adamo-v4
28 pearls-transients-v4
325 rubies-egs51-v4
439 rubies-egs52-v4
450 rubies-egs53-v4
406 rubies-egs61-v4
497 rubies-egs62-v4
479 rubies-egs63-v4
516 rubies-uds1-v4
478 rubies-uds2-v4
433 rubies-uds21-v4
437 rubies-uds22-v4
405 rubies-uds23-v4
496 rubies-uds3-v4
443 rubies-uds31-v4
512 rubies-uds32-v4
491 rubies-uds33-v4
496 rubies-uds41-v4
471 rubies-uds42-v4
465 rubies-uds43-v4
225 rxj2129-ddt-v4
86 smacs0723-ero-v4
123 snh0pe-v4
78 spt0615-v4
92 stark-a1703-v4
147 stark-rxcj2248-v4
62 suspense-kriek-v4
192 ulas-j1120-gto-v4
154 uncover-61-v4
188 uncover-62-v4
559 uncover-v4
62 valentino-cosmos02-v4
46 valentino-cosmos03-v4
67 valentino-cosmos04-v4
184 valentino-egs-v4
47 valentino-obs08-v4
99 valentino-obs10-v4
68 valentino-obs12-v4
58 valentino-obs15-v4
55 valentino-obs19-v4
106 weisz-leoa-v4
77 weisz-tucana-v4
225 westerlund2-imf-v4
454 whl0137-v4
<grizli.utils.Unique at 0x307cc9130>
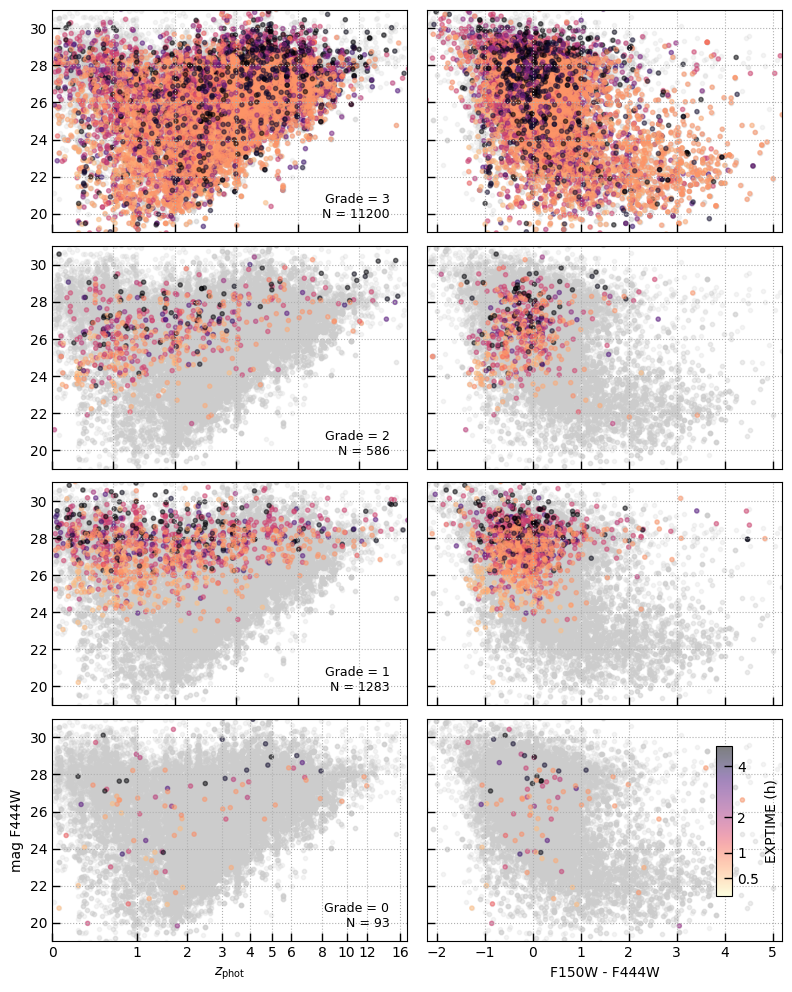
Source counts by grade
Show magnitude, color, redshift distribution as a function of the visual classification grade:
3: Robust redshift from one or more emission absorption features2: Ambiguous continuum features, perhaps only one line or low confidence lines1: No clear features in the spetrum to constrain the redshift0: Spectrum suffers some data quality issue and should-1: (Spectrum did not have grade from visual inspection)
fig, axes = plt.subplots(4,2,figsize=(8,10), sharex=False, sharey=True)
colors = {0: 'magenta', 1: '0.5', 2: 'coral', 3: 'olive'}
# sub = is_rubies
sub = tab['ra'] > 0
sub = sub & True
sub &= tab['z_phot'].filled(-1) > 0
sub &= tab['grating'] == 'PRISM'
un = utils.Unique(tab[sub]['grade'].filled(-1))
blue = -2.5*np.log10(tab['phot_f150w_tot_1'] / tab['phot_f444w_tot_1'])
for i, c in enumerate([3,2,1,0]):
kws = dict(
c = np.sqrt(tab[sub][un[c]]['exptime']), vmin=900**0.5, vmax=(5*3600)**0.5, cmap='magma_r',
# c = 'magenta',
alpha=0.5,
label=f'Grade = {c}',
)
ax = axes[i][1]
ax.scatter(blue,
23.9 - 2.5*np.log10(tab['phot_f444w_tot_1']),
c='0.8',
alpha=0.2,
label=f'Grade = {c}',
)
sc = ax.scatter(blue[sub][un[c]],
23.9 - 2.5*np.log10(tab[sub]['phot_f444w_tot_1'])[un[c]],
**kws,
)
ax.grid()
if i < 3:
ax.set_xticklabels([])
ax.set_xlim(-2.2, 5.2)
ax = axes[i][0]
ax.scatter(np.log(1+tab['z_phot']),
23.9 - 2.5*np.log10(tab['phot_f444w_tot_1']),
c='0.8',
alpha=0.2,
label=f'Grade = {c}' + '\n' + f'N = {un[c].sum()}',
)
ax.scatter(np.log(1+tab[sub]['z_phot'][un[c]]),
23.9 - 2.5*np.log10(tab[sub]['phot_f444w_tot_1'])[un[c]],
**kws,
)
ax.grid()
ax.text(
0.95, 0.05,
# f'Grade = {c}',
f'Grade = {c}' + '\n' + f'N = {un[c].sum()}',
ha='right', va='bottom', fontsize=9, transform=ax.transAxes)
if i < 3:
ax.set_xticklabels([])
xt = [0, 1, 2, 3, 4, 5, 6, 8, 10, 12, 16]
ax.set_xlim(0, np.log(1+17))
ax.set_ylabel('mag F444W')
cax = fig.add_axes
cax = fig.add_axes((0.9, 0.1, 0.02, 0.15))
cb = plt.colorbar(sc, cax=cax, orientation='vertical')
ct = [0.5, 1, 2, 4]
cb.set_ticks(np.sqrt(np.array(ct)*3600))
cb.set_ticklabels(ct)
cb.set_label('EXPTIME (h)')
ax.set_ylim(19, 31)
ax.set_xticks(np.log(1+np.array(xt)))
ax.set_xticklabels(xt)
ax.set_xlabel(r'$z_\mathrm{phot}$')
ax = axes[3][1]
ax.set_xlabel('F150W - F444W')
# ax.legend()
fig.tight_layout(pad=1)
N value
==== ==========
4799 -1
114 0
1697 1
815 2
13703 3
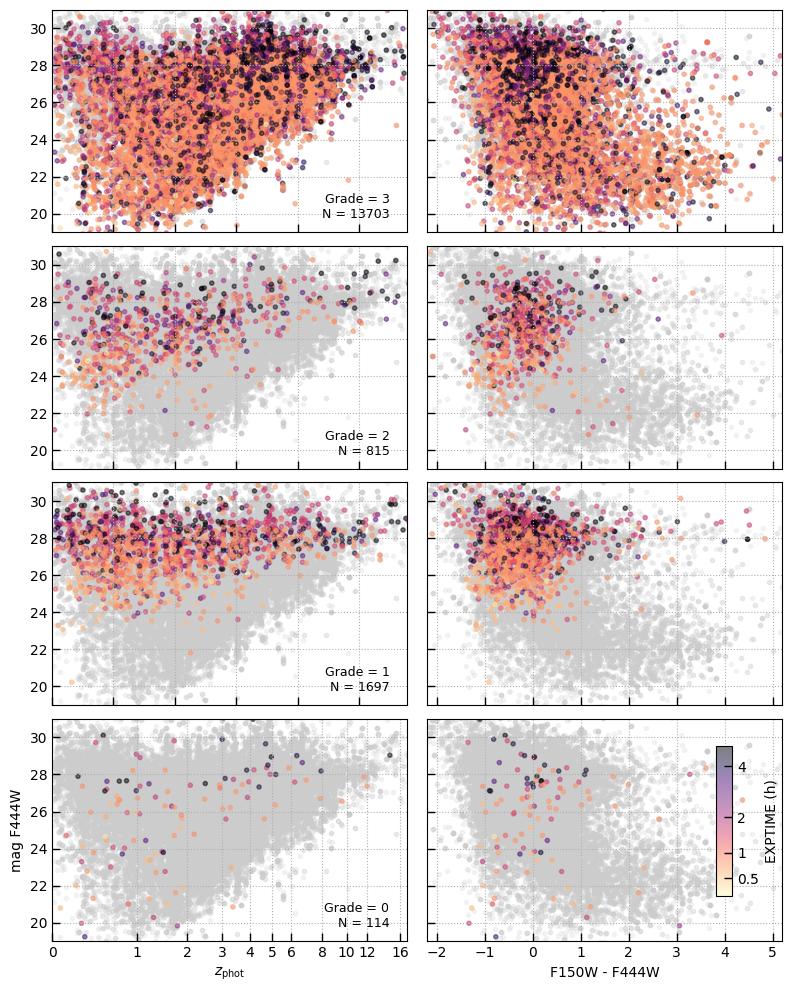
PRISM sample for comparision
is_prism = (tab['grating'] == 'PRISM')
sample = is_prism & (tab['grade'] == 3)
sample &= (tab['z_best'] < 7)
sample &= (tab['zrf'] > 0) & (tab['z_best'] > 0)
sample &= tab['rest_153_frac'] > 0.8
sample &= tab['rest_154_frac'] > 0.8
sample &= tab['rest_155_frac'] > 0.8
sample.sum()
15468
# Compare the redshift where the emission line fits were performed to the
# "best" redshift calculated for discrete unique sources
_ = eazy.utils.zphot_zspec(tab['zrf'][is_prism], tab['z_best'][is_prism], zmax=8)
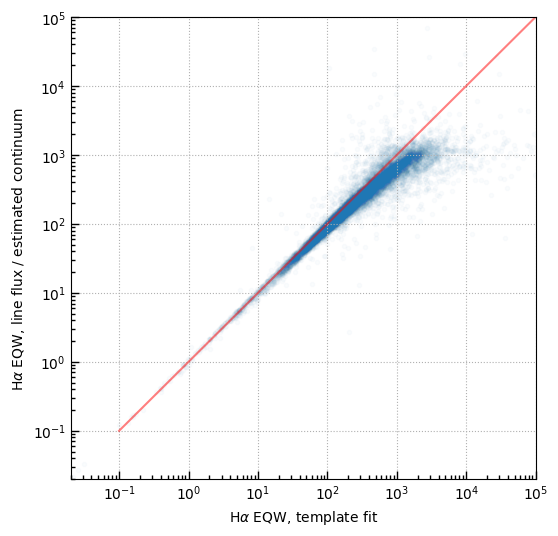
Interpolate Halpha EQW from nearby filters
import eazy.filters
RES = eazy.filters.FilterFile()
fb, fr = 415, 416
#fb, fr = 155, 416
wb = RES[fb].pivot
wr = RES[fr].pivot
flamb = (1*u.microJansky).to(u.erg/u.second/u.cm**2/u.Angstrom, equivalencies=u.spectral_density(wb*u.Angstrom))
flamr = (1*u.microJansky).to(u.erg/u.second/u.cm**2/u.Angstrom, equivalencies=u.spectral_density(wr*u.Angstrom))
whtb = (1 - np.abs(wb - 6564.)/(wr-wb))# *flamb
whtr = (1 - np.abs(wr - 6564.)/(wr-wb))# *flamr
interp_flux = tab[f'rest_{fb}_flux']*whtb*flamb + tab[f'rest_{fr}_flux']*whtr*flamr
eqw = ((tab['line_ha_nii']*1.e-20*u.erg/u.second/u.cm**2 / (interp_flux / (1+tab['zline'])**1))).value
plt.scatter(
(np.maximum(tab['eqw_ha_nii'], -100) / (1+tab['zline'])**1)[sample],
eqw[sample], alpha=0.02
)
plt.plot([0.1, 1e7], [0.1, 1e7], color='r', alpha=0.5)
plt.loglog()
plt.grid()
plt.xlim(0.02, 1.e5); plt.ylim(0.02, 1.e5)
plt.xlabel(r'H$\alpha$ EQW, template fit')
plt.ylabel(r'H$\alpha$ EQW, line flux / estimated continuum')
if 1:
print('Use interpolated EQW')
eqw_lim = np.maximum(tab['line_ha_nii'], tab['line_ha_nii_err']*2) * 1.e-20*u.erg/u.second/u.cm**2 / (interp_flux / (1+tab['zline']))
is_eqw_lim = tab['line_ha_nii_err']*2 > tab['line_ha_nii']
eqw[is_eqw_lim] = eqw_lim.value[is_eqw_lim]
tab['ha_eqw_with_limits'] = eqw
tab['ha_eqw_is_limit'] = is_eqw_lim
Use interpolated EQW
Stellar population properties
- Rest-frame colors
- Stellar masses
- …
UV = -2.5*np.log10(tab['phot_restU'] / tab['phot_restV'])
VJ = -2.5*np.log10(tab['phot_restV'] / tab['phot_restJ'])
UVs = -2.5*np.log10(tab['rest_153_flux'] / tab['rest_155_flux'])
BVs = -2.5*np.log10(tab['rest_154_flux'] / tab['rest_155_flux'])
VJs = -2.5*np.log10(tab['rest_155_flux'] / tab['rest_161_flux'])
eBVs = 2.5/np.log(10) * np.sqrt(
(tab['rest_154_full_err'] / tab['rest_154_flux'])**2
+ (tab['rest_155_full_err'] / tab['rest_155_flux'])**2
)
ugs = -2.5*np.log10(tab['rest_414_flux'] / tab['rest_415_flux'])
gis = -2.5*np.log10(tab['rest_415_flux'] / tab['rest_416_flux'])
ok_BVs = (tab['rest_154_frac'] > 0.8) & (tab['rest_155_frac'] > 0.8)
ok_gis = (tab['rest_415_frac'] > 0.8) & (tab['rest_416_frac'] > 0.8)
ok_BVs &= eBVs < 0.1
dL = WMAP9.luminosity_distance(tab['zrf']).to('cm')
rest_fV = (tab['rest_155_flux']*u.microJansky).to(
u.erg/u.second/u.cm**2/u.Angstrom,
equivalencies=u.spectral_density(5500.*(1+tab['zrf'])*u.Angstrom)
)
rest_fi = (tab['rest_416_flux']*u.microJansky).to(
u.erg/u.second/u.cm**2/u.Angstrom,
equivalencies=u.spectral_density(RES[416].pivot * (1+tab['zrf'])*u.Angstrom)
)
LV = (rest_fV * 5500. * u.Angstrom * (1 + tab['zrf']) * 4 * np.pi * dL**2).to(u.Lsun)
Li = (rest_fi * RES[416].pivot * u.Angstrom * (1 + tab['zrf']) * 4 * np.pi * dL**2).to(u.Lsun)
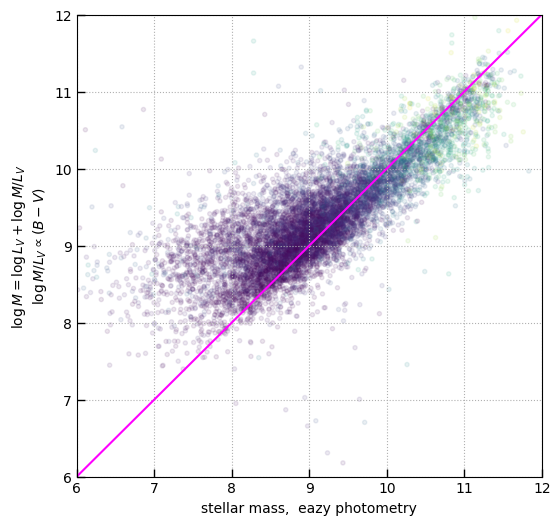
# Crude M/Lv ~ B-V from Taylor et al. 2009 for getting a quick stellar mass from the spectrum
log_MLv = -0.734 + 1.404 * (BVs + 0.084)
MassV = log_MLv + np.log10(LV.value)
tab['Mass'] = MassV
tab['Mass'].format = '.2f'
tab['ok_Mass'] = ok_BVs
plt.scatter(
np.log10(tab['phot_mass'][sample & ok_BVs]),
MassV[sample & ok_BVs],
alpha=0.1,
c=tab['phot_Av'][sample & ok_BVs]
)
plt.plot([5, 12], [5, 12], color='magenta')
plt.grid()
plt.xlim(6, 12)
plt.ylim(6, 12)
plt.xlabel('stellar mass, eazy photometry')
plt.ylabel(r'$\log M = \log L_V + \log M/L_V$' + '\n' + r'$\log M/L_V \propto (B-V)$')
Text(0, 0.5, '$\\log M = \\log L_V + \\log M/L_V$\n$\\log M/L_V \\propto (B-V)$')
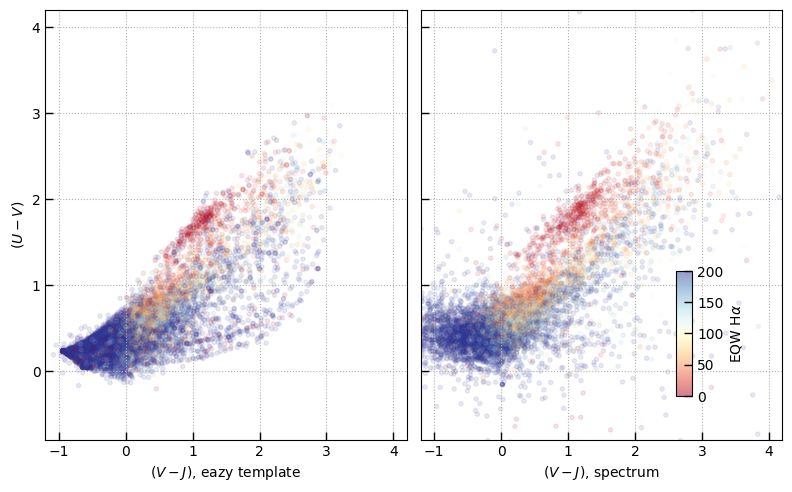
Compare rest-frame colors
The table includes rest-frame bandpass flux densities 1) estimated from the broad-band photometry (at the photo-z) and 2) integrated directly through the spectra at the measured redshift.
The colors derived from the grizli/DJA photometry are those of the best-fit photo-z template combination, not a noisy interpolation, so they can show banding effects resulting from the discrete combination of templates.
fig, axes = plt.subplots(1,2,figsize=(8,5), sharex=True, sharey=True)
axes[0].scatter(
VJ[sample], UV[sample], alpha=0.1,
c=tab['ha_eqw_with_limits'][sample], vmin=0, vmax=200, cmap='RdYlBu'
)
axes[0].set_xlabel(r'$(V-J)$' + ', eazy template')
axes[0].set_ylabel(r'$(U-V)$')
axes[1].scatter(
VJs[sample], UVs[sample], alpha=0.1,
c=tab['ha_eqw_with_limits'][sample], vmin=0, vmax=200, cmap='RdYlBu'
)
sc = axes[1].scatter(
VJs[sample][:1], UVs[sample][:1], alpha=0.5,
c=tab['ha_eqw_with_limits'][sample][:1], vmin=0, vmax=200, cmap='RdYlBu'
)
axes[1].set_xlabel(r'$(V-J)$' + ', spectrum')
for ax in axes:
ax.set_xlim(-1.2, 4.2)
ax.set_ylim(-0.8, 4.2)
ax.grid()
cax = fig.add_axes((0.85, 0.2, 0.02, 0.25))
cb = plt.colorbar(sc, cax=cax, orientation='vertical')
cb.set_label(r'EQW H$\alpha$')
fig.tight_layout(pad=1)
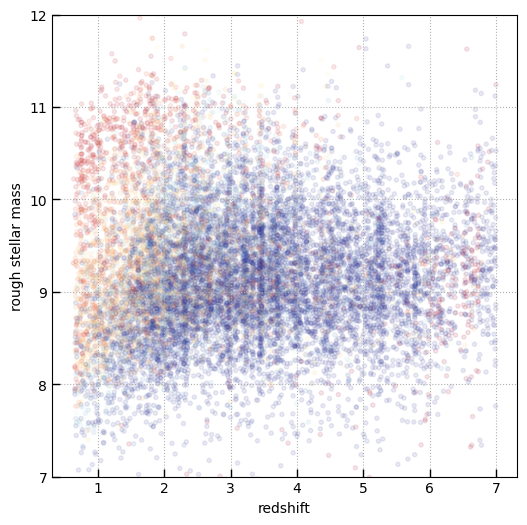
plt.scatter(
tab['z_best'][sample],
# np.log10(tab['phot_mass'])[sample],
tab['Mass'][sample],
alpha=0.1,
c=tab['ha_eqw_with_limits'][sample], vmin=0, vmax=200, cmap='RdYlBu'
)
plt.ylim(7, 12)
plt.grid()
plt.xlabel('redshift')
plt.ylabel('rough stellar mass')
Text(0, 0.5, 'rough stellar mass')
Make a table showing thumbnail and spectrum previews of a selected subsample
Here make a “massive galaxies” subsample with
grating=PRISMz > 3log M > 10.5- Spectrum covers rest-frame $B-V$
The preview table shows the first 32 of these, which actually tend to be quasars / LRDs where the stellar mass is likely incorrect….
massive = sample & (tab['z_best'] > 3.) & (MassV > 10.5) & (tab['grating'] == 'PRISM') & ok_BVs
if 0:
tab['root','file','z_best','Mass','ha_eqw_with_limits','Thumb','Slit_Thumb','Spectrum_fnu', 'Spectrum_flam'][massive].write_sortable_html(
'/tmp/massive.html',
max_lines=1000,
localhost=False,
)
print(f"massive test sample: {massive.sum()}")
massive test sample: 235
from IPython.display import display, Markdown, Latex
so = np.argsort(tab['Mass'][massive])[::-1]
so = so[:32]
df = tab['root','file','z_best','Mass','ha_eqw_with_limits','Thumb','Slit_Thumb','Spectrum_fnu', 'Spectrum_flam'][massive][so].to_pandas()
display(Markdown(df.to_markdown()))
| root | file | z_best | Mass | ha_eqw_with_limits | Thumb | Slit_Thumb | Spectrum_fnu | Spectrum_flam | |
|---|---|---|---|---|---|---|---|---|---|
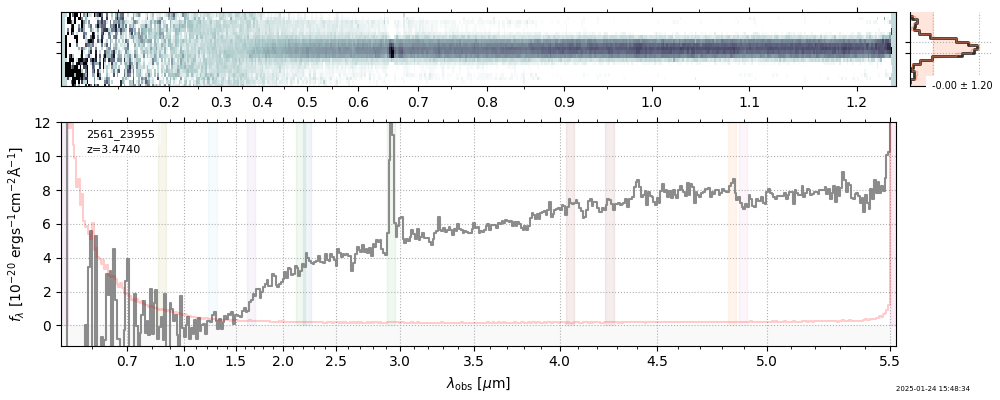
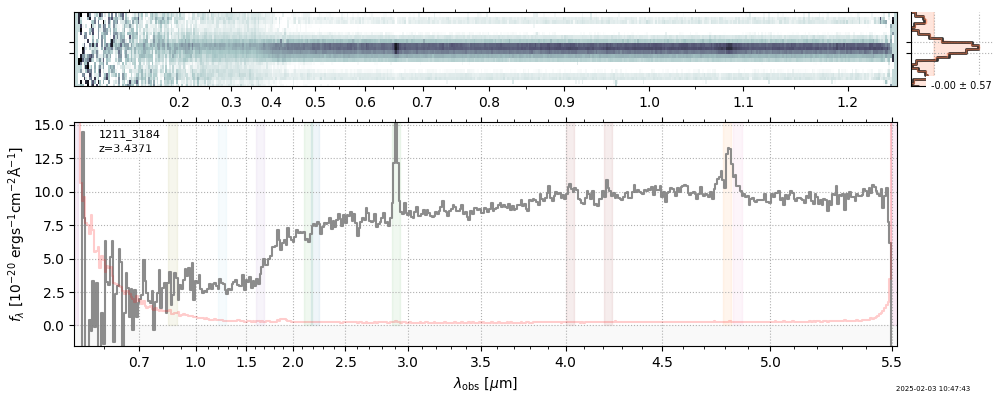
| 0 | j0910-wang-v4 | j0910-wang-v4_prism-clear_2028_12910.spec.fits | 6.62142 | 12.0515 | 54.4902 |  |
 |
||
| 1 | rubies-uds23-v4 | rubies-uds23-v4_prism-clear_4233_166691.spec.fits | 4.06673 | 11.9254 | 11.4969 |  |
 |
||
| 2 | uncover-61-v4 | uncover-61-v4_prism-clear_2561_13416.spec.fits | 4.02262 | 11.7896 | 114.235 | 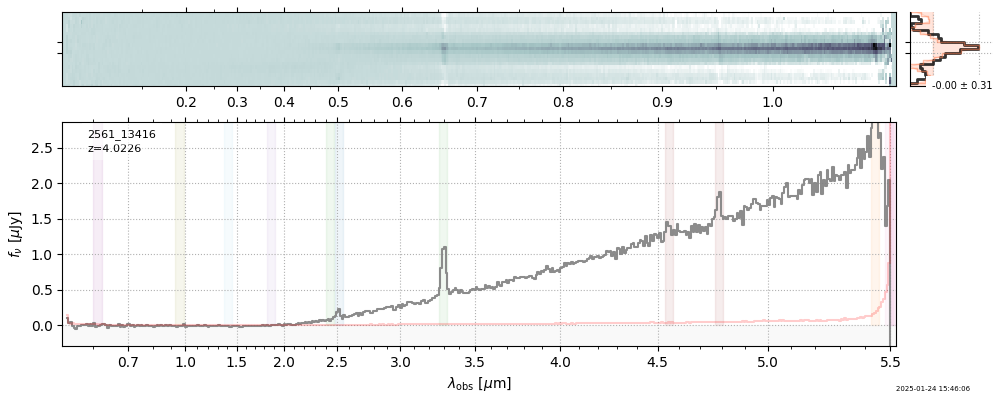 |
 |
||
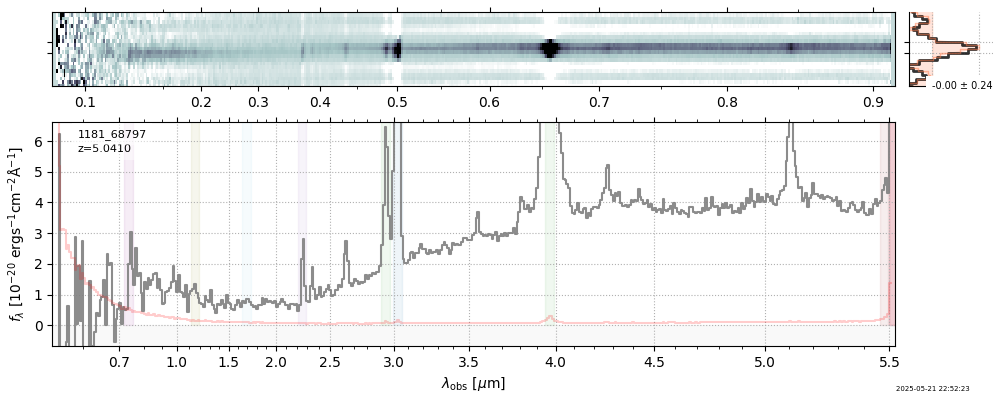
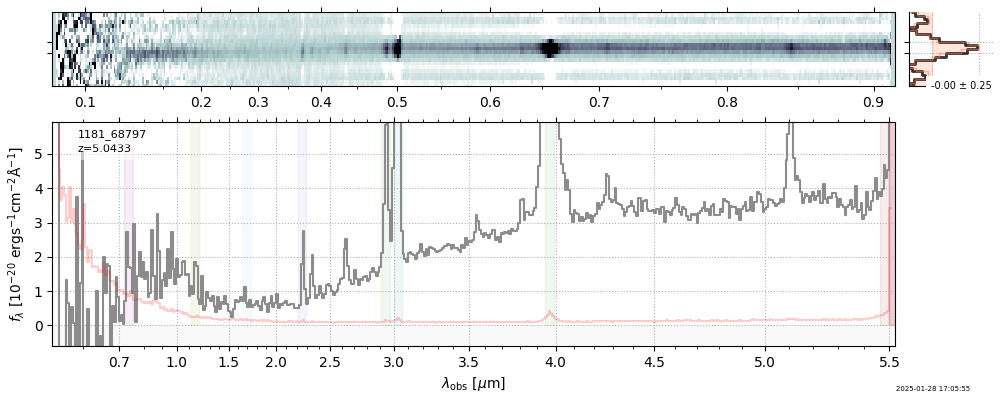
| 3 | jades-gdn198-v4 | jades-gdn198-v4_prism-clear_1181_68797.spec.fits | 5.0398 | 11.7363 | 1029.12 | 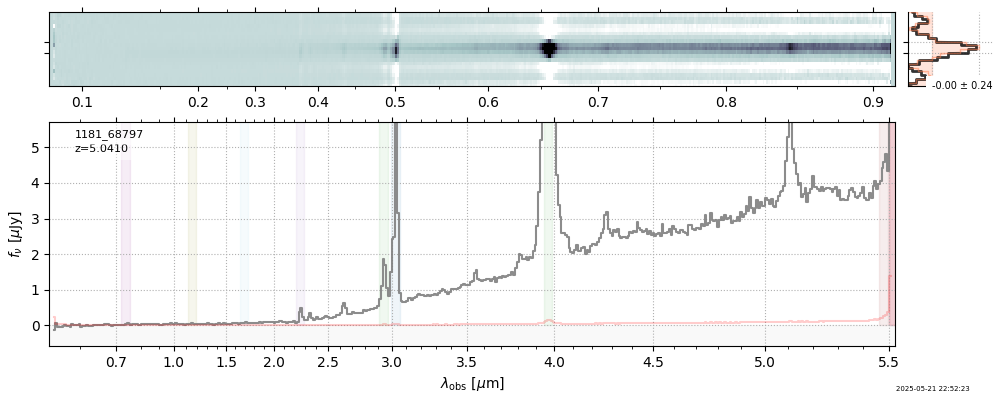 |
 |
||
| 4 | rubies-egs52-v4 | rubies-egs52-v4_prism-clear_4233_9809.spec.fits | 5.68123 | 11.6563 | 197.61 |  |
 |
||
| 5 | jades-gdn-v4 | jades-gdn-v4_prism-clear_1181_68797.spec.fits | 5.0398 | 11.6303 | 1054.65 | 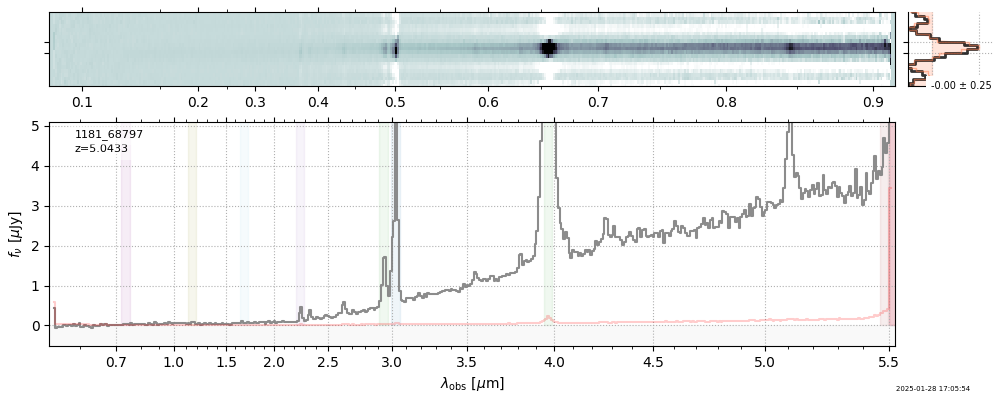 |
 |
||
| 6 | rubies-uds22-v4 | rubies-uds22-v4_prism-clear_4233_114988.spec.fits | 4.36474 | 11.5051 | 94.5801 | 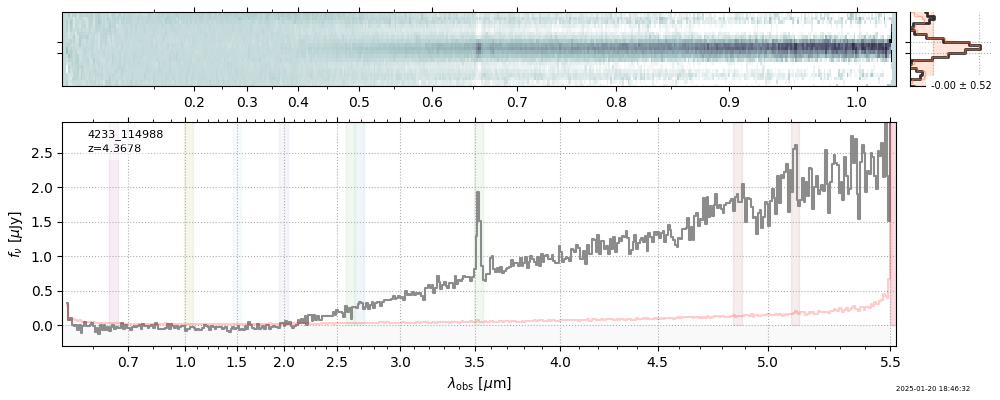 |
 |
||
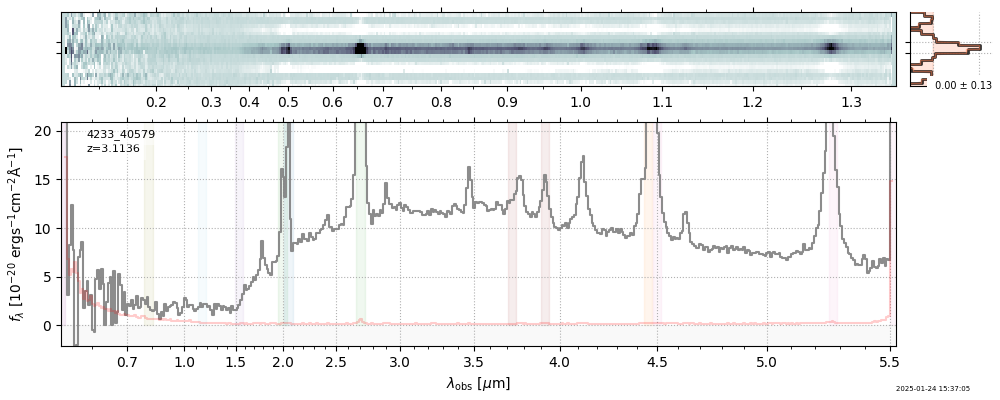
| 7 | rubies-uds1-v4 | rubies-uds1-v4_prism-clear_4233_40579.spec.fits | 3.10671 | 11.4453 | 1206.13 | 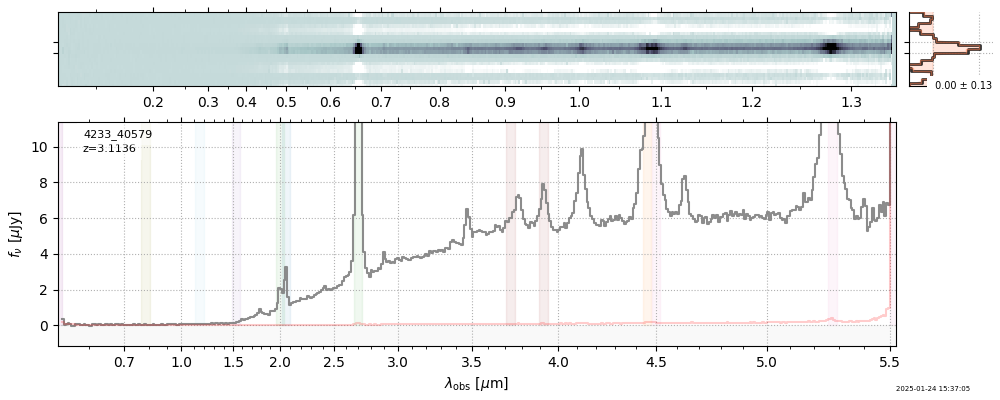 |
 |
||
| 8 | gto-wide-uds13-v4 | gto-wide-uds13-v4_prism-clear_1215_1472.spec.fits | 4.55596 | 11.4348 | 14.736 |  |
 |
||
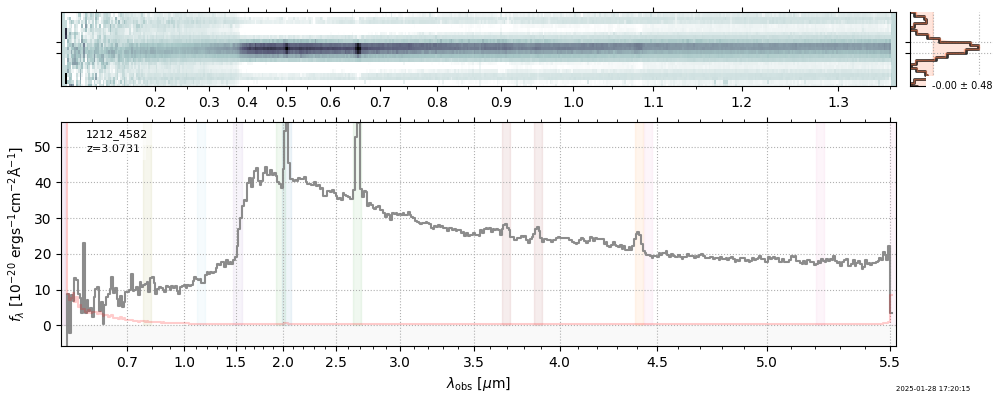
| 9 | jades-gds-w05-v4 | jades-gds-w05-v4_prism-clear_1212_4582.spec.fits | 3.06306 | 11.3624 | 79.7105 | 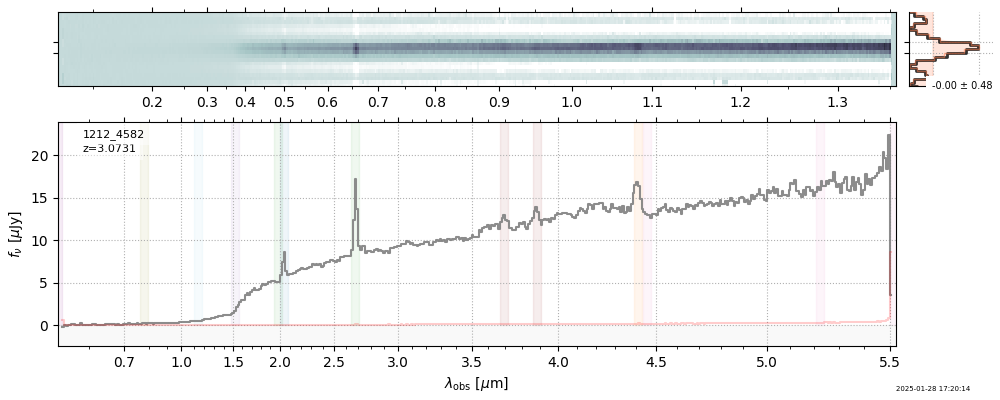 |
 |
||
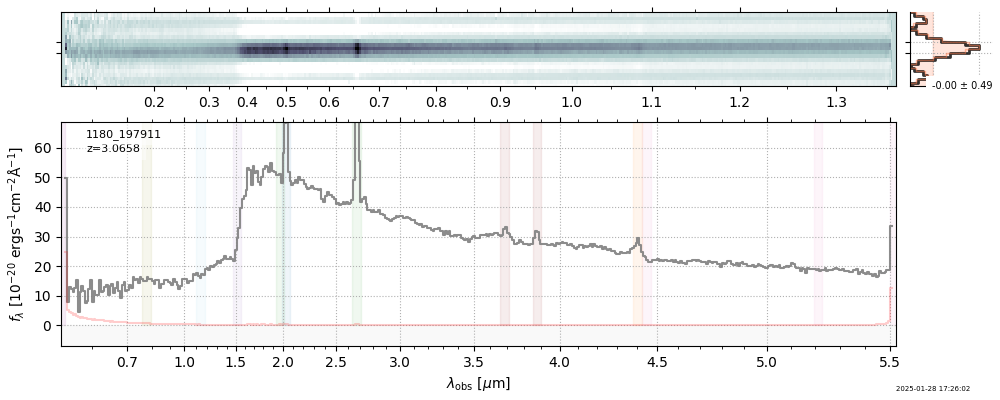
| 10 | jades-gds-wide3-v4 | jades-gds-wide3-v4_prism-clear_1180_197911.spec.fits | 3.06306 | 11.3613 | 80.9861 | 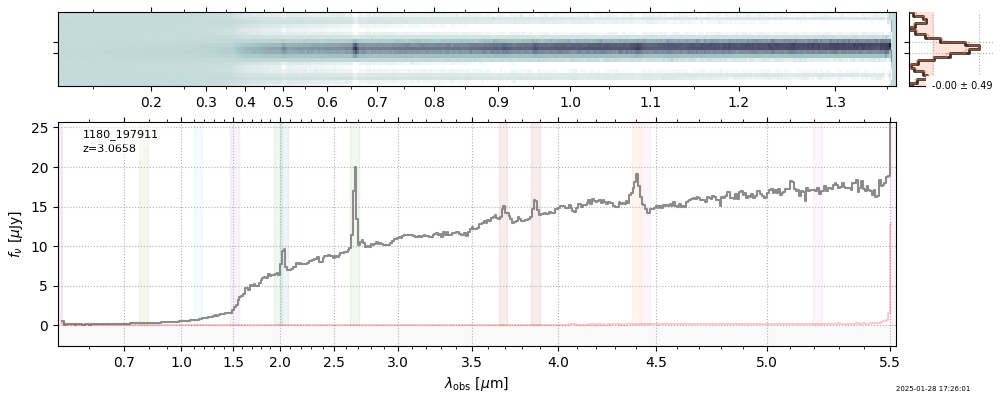 |
 |
||
| 11 | uncover-v4 | uncover-v4_prism-clear_2561_45924.spec.fits | 4.4673 | 11.3522 | 104.309 | 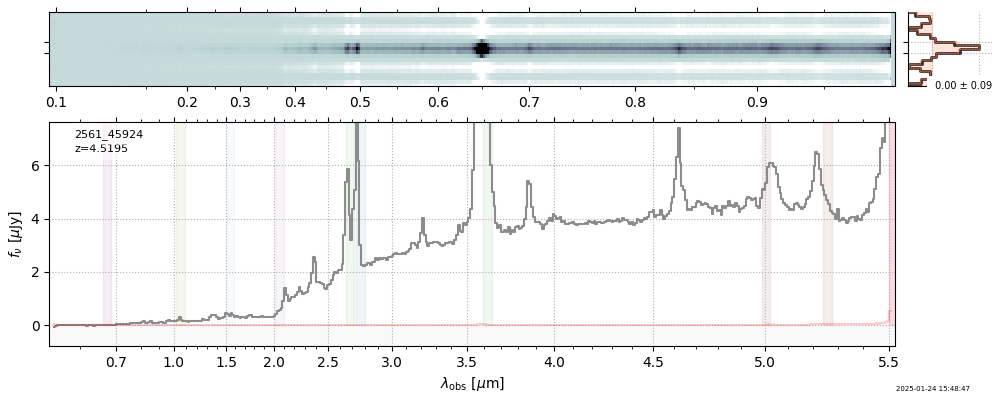 |
 |
||
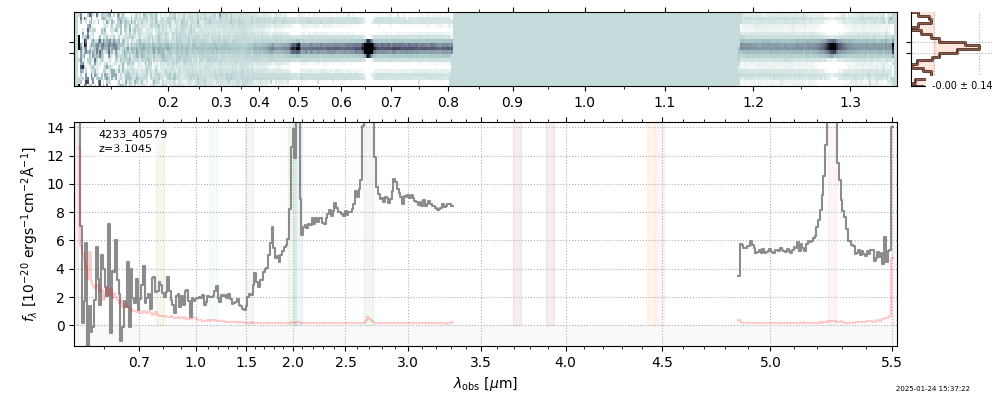
| 12 | rubies-uds2-v4 | rubies-uds2-v4_prism-clear_4233_40579.spec.fits | 3.10671 | 11.3515 | 1369.53 | 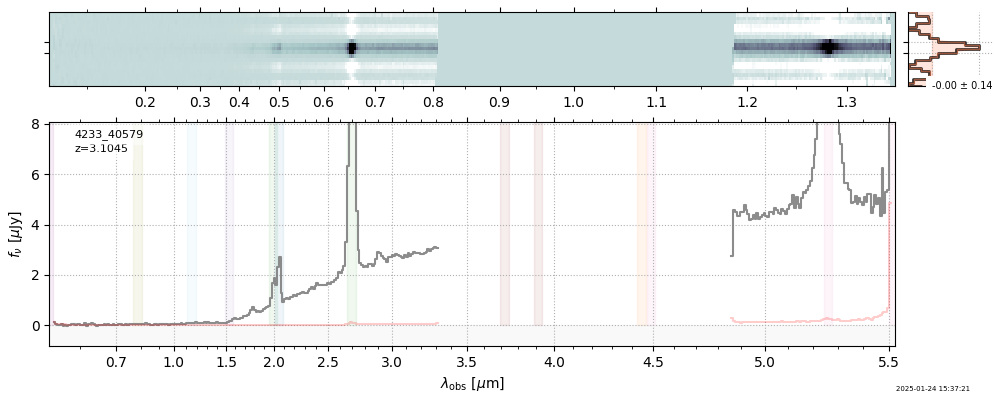 |
 |
||
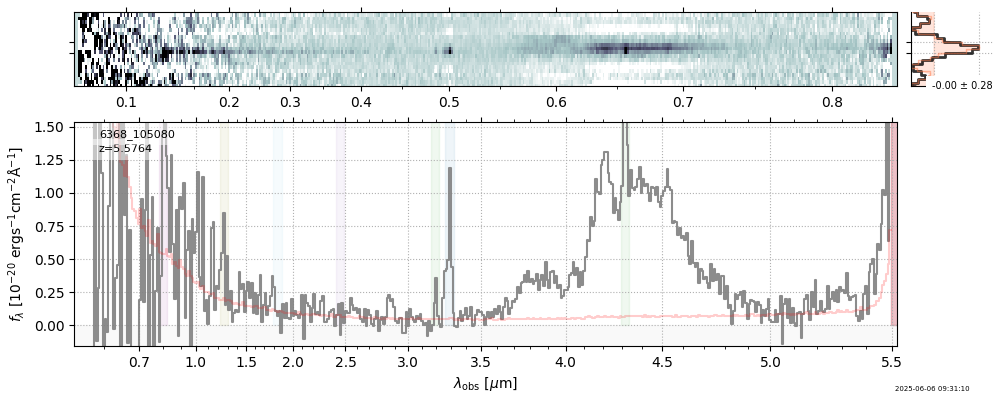
| 13 | capers-cos07-v4 | capers-cos07-v4_prism-clear_6368_105080.spec.fits | 5.58011 | 11.3182 | 138.384 | 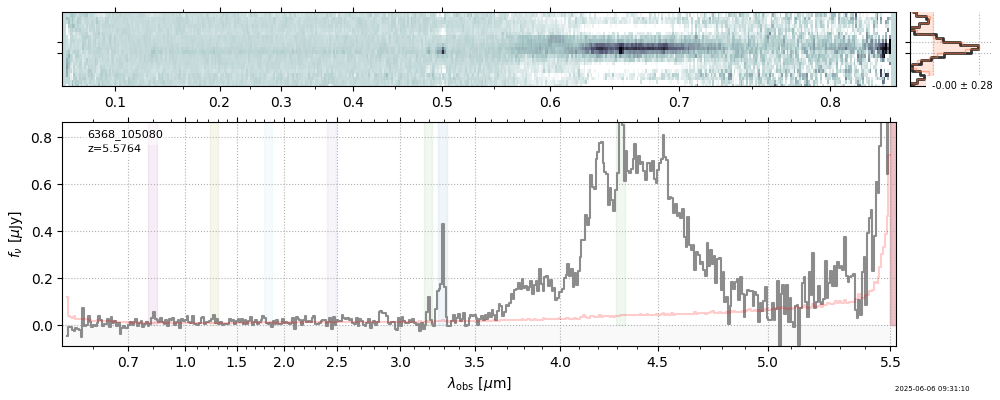 |
 |
||
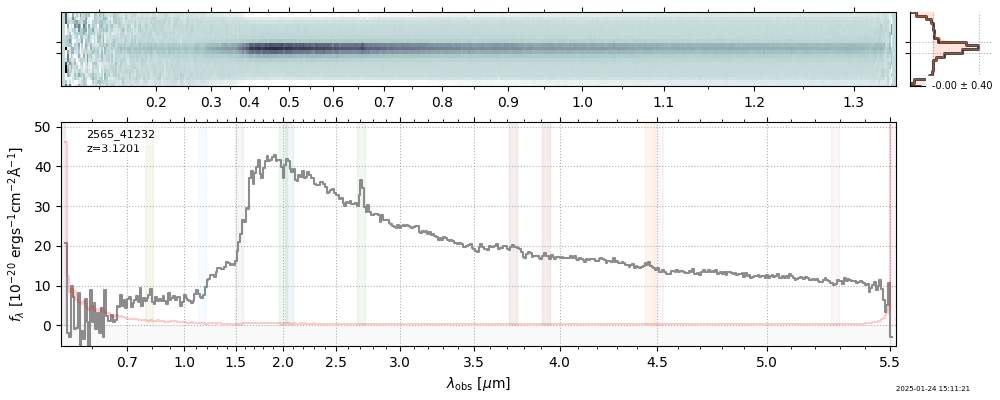
| 14 | glazebrook-v4 | glazebrook-v4_prism-clear_2565_41232.spec.fits | 3.12015 | 11.2815 | 17.5962 | 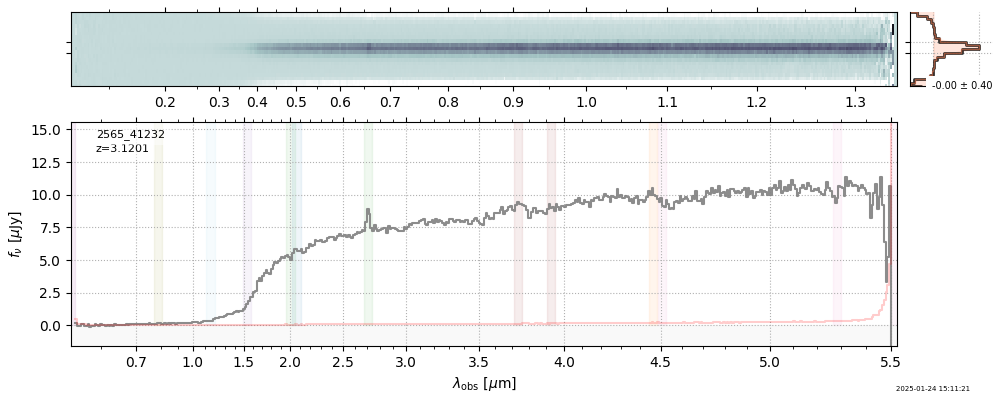 |
 |
||
| 15 | nexus-obs3-v4 | nexus-obs3-v4_prism-clear_5105_23192.spec.fits | 4.52105 | 11.2767 | 1058.6 | 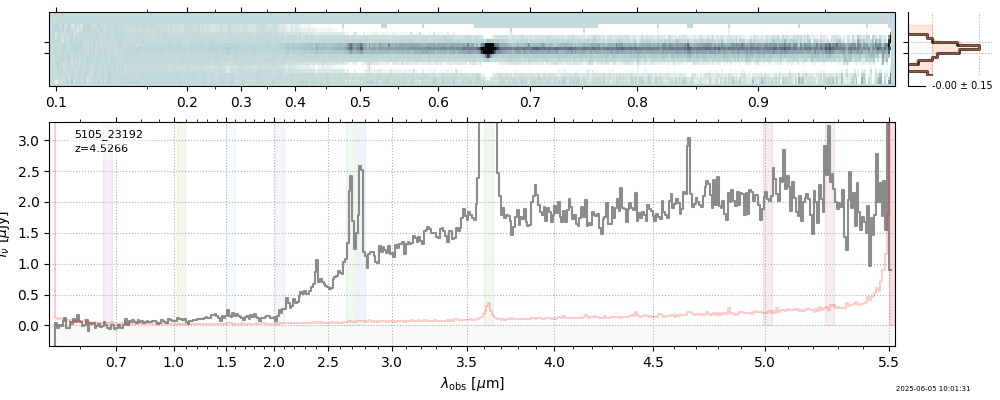 |
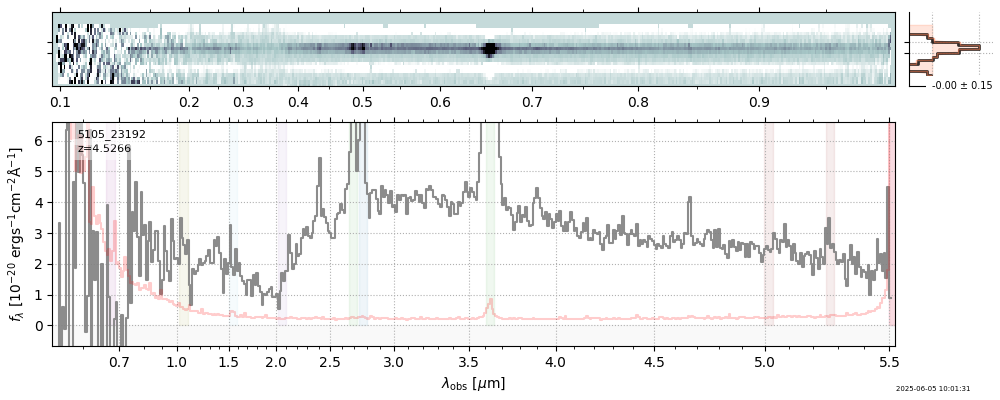 |
||
| 16 | capers-egs65-v4 | capers-egs65-v4_prism-clear_6368_27615.spec.fits | 5.68123 | 11.2409 | 241.543 | 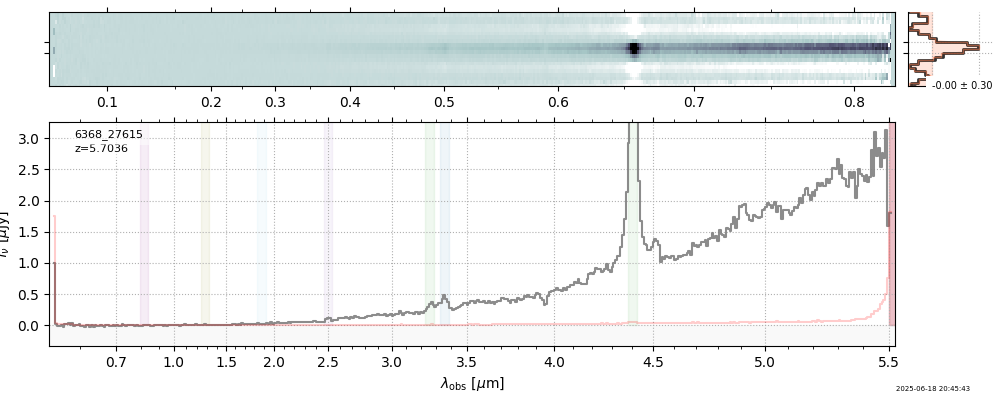 |
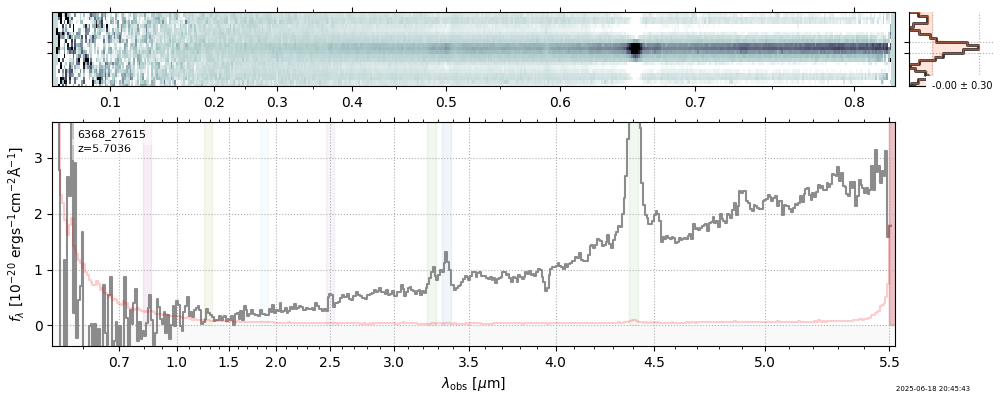 |
||
| 17 | uncover-v4 | uncover-v4_prism-clear_2561_45092.spec.fits | 3.46158 | 11.2391 | 98.5119 | 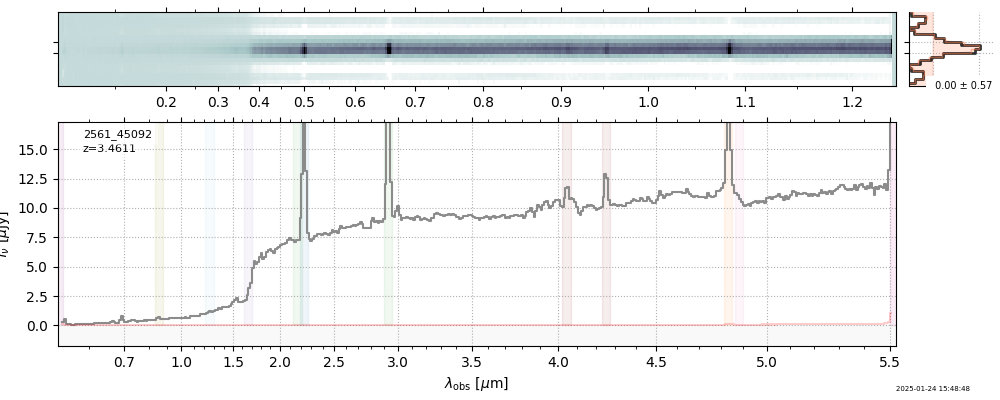 |
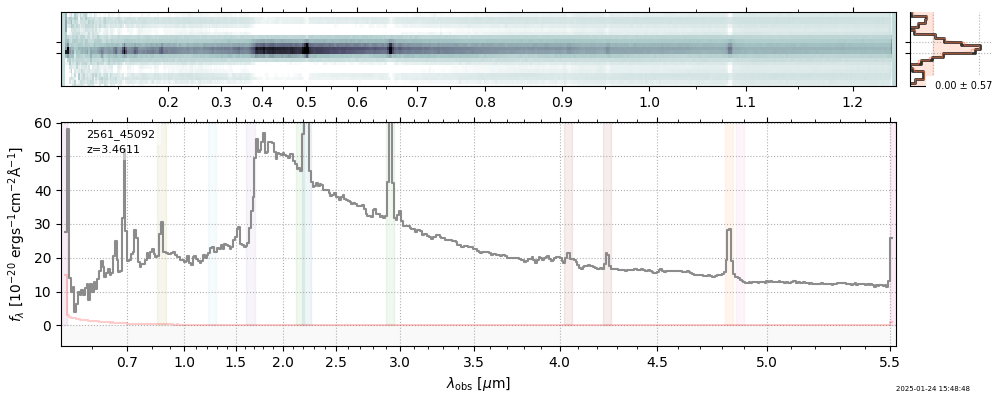 |
||
| 18 | uncover-62-v4 | uncover-62-v4_prism-clear_2561_57618.spec.fits | 3.46158 | 11.2264 | 82.8891 | 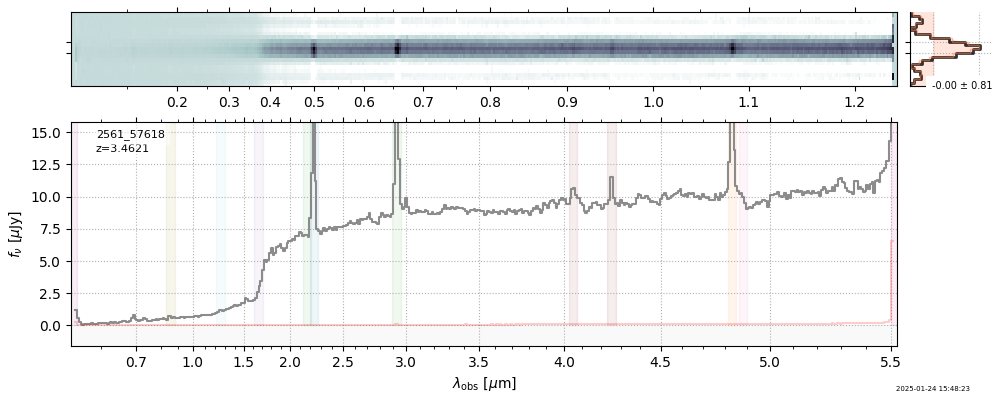 |
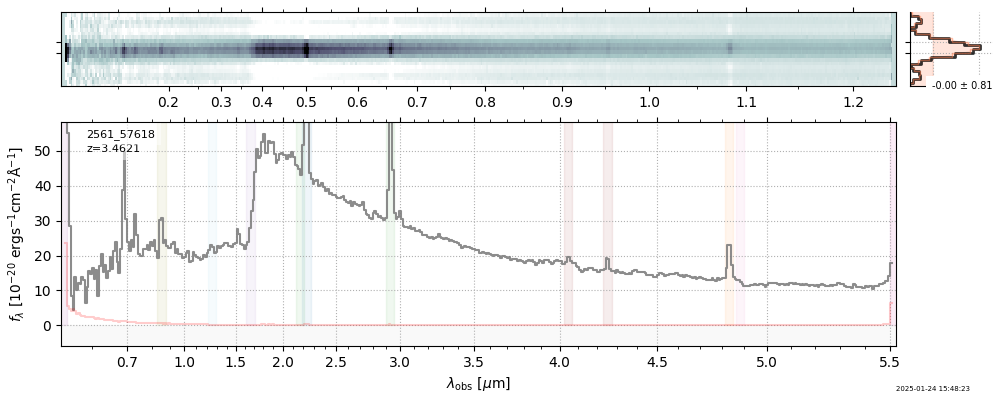 |
||
| 19 | uncover-62-v4 | uncover-62-v4_prism-clear_2561_58453.spec.fits | 4.4673 | 11.2252 | 969.994 | 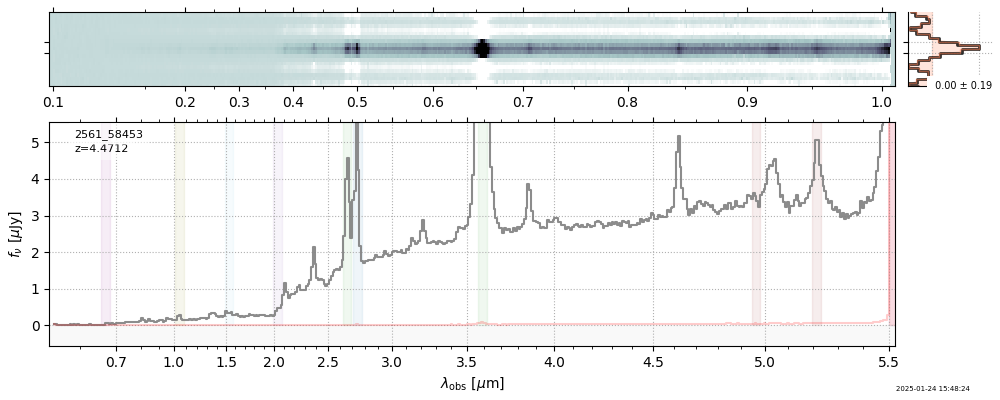 |
 |
||
| 20 | gds-barrufet-s67-v4 | gds-barrufet-s67-v4_prism-clear_2198_1260.spec.fits | 4.4319 | 11.2251 | 72.7017 |  |
 |
||
| 21 | glazebrook-cos-obs3-v4 | glazebrook-cos-obs3-v4_prism-clear_2565_20115.spec.fits | 3.71293 | 11.2231 | 2.61723 |  |
 |
||
| 22 | glazebrook-v4 | glazebrook-v4_prism-clear_2565_12629.spec.fits | 3.19399 | 11.2039 | 5.4745 |  |
 |
||
| 23 | rubies-egs63-v4 | rubies-egs63-v4_prism-clear_4233_49140.spec.fits | 6.68847 | 11.1816 | 657.49 |  |
 |
||
| 24 | jades-gds-w08-v4 | jades-gds-w08-v4_prism-clear_1212_792.spec.fits | 3.67516 | 11.173 | 341.192 |  |
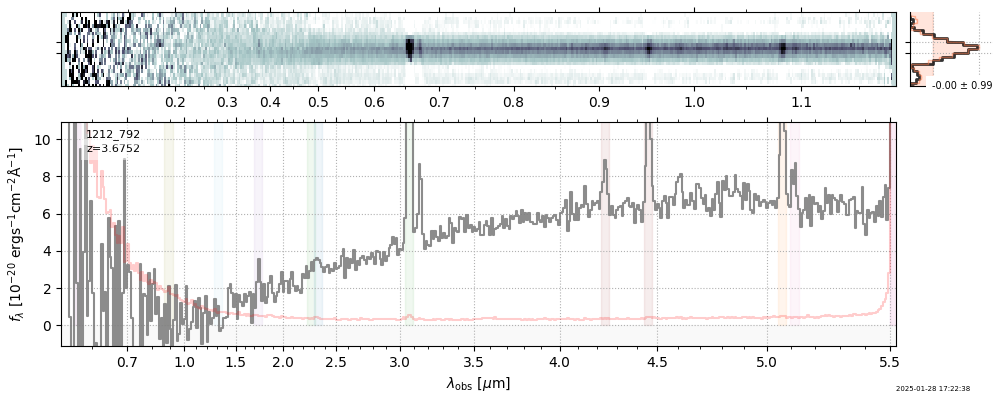 |
||
| 25 | rubies-uds42-v4 | rubies-uds42-v4_prism-clear_4233_807469.spec.fits | 6.77538 | 11.1462 | 4951.05 | 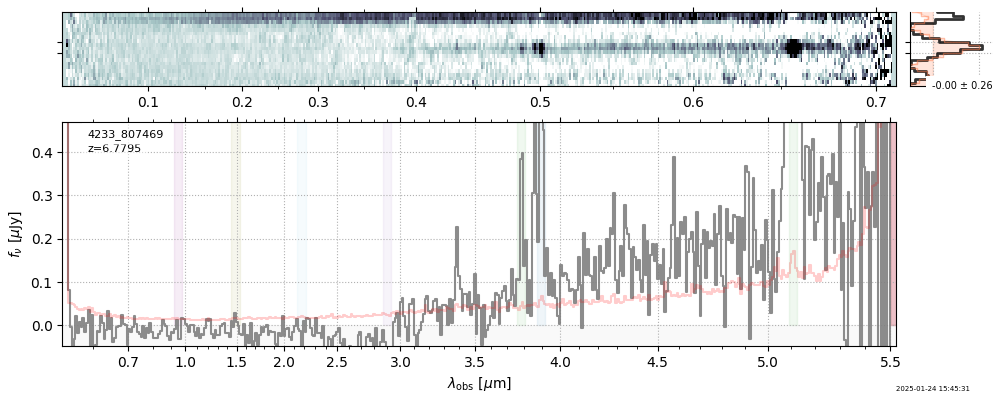 |
 |
||
| 26 | rubies-egs53-v4 | rubies-egs53-v4_prism-clear_4233_25712.spec.fits | 3.89179 | 11.1396 | 120.361 | 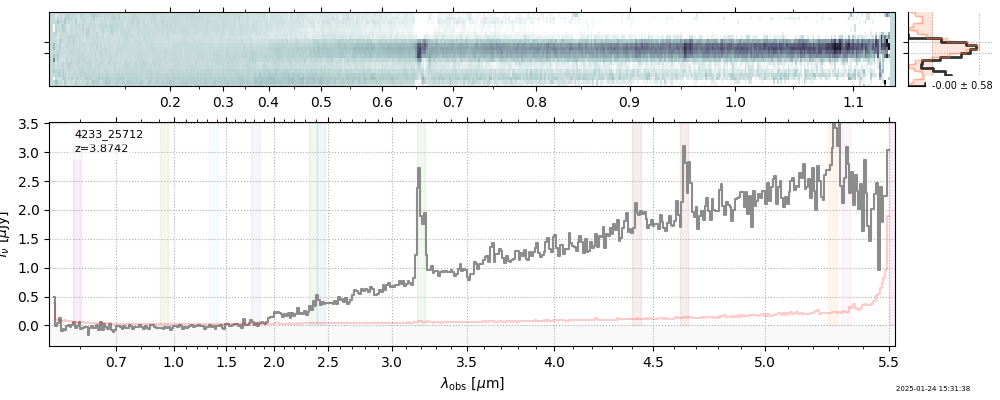 |
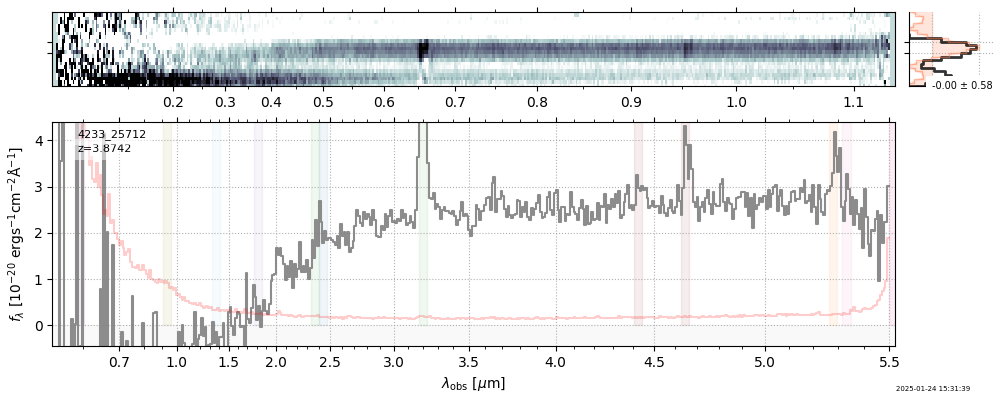 |
||
| 27 | uncover-v4 | uncover-v4_prism-clear_2561_23955.spec.fits | 3.47275 | 11.1197 | 131.122 | 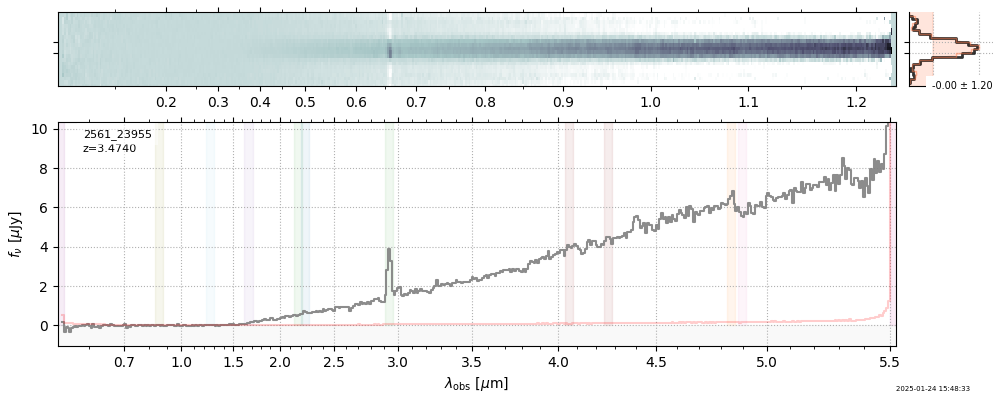 |
 |
||
| 28 | goodsn-wide66-v4 | goodsn-wide66-v4_prism-clear_1211_3184.spec.fits | 3.4402 | 11.1181 | 59.2013 | 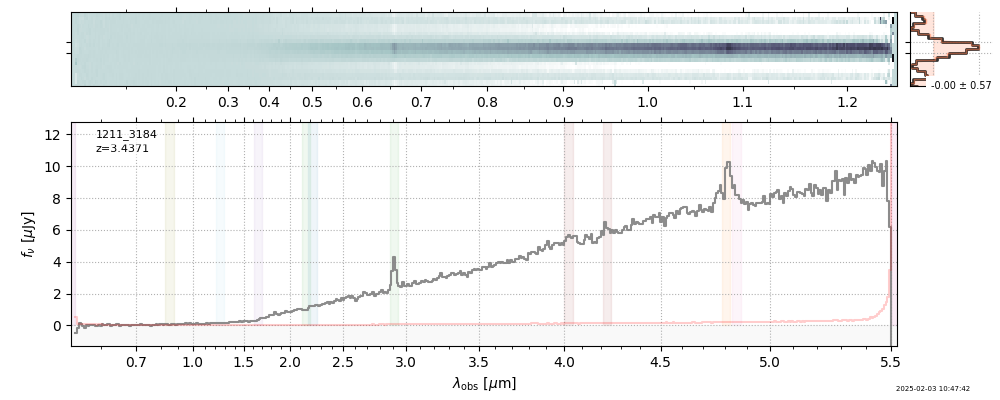 |
 |
||
| 29 | rubies-egs61-v4 | rubies-egs61-v4_prism-clear_4233_55604.spec.fits | 6.98435 | 11.1174 | 2762.98 |  |
 |
||
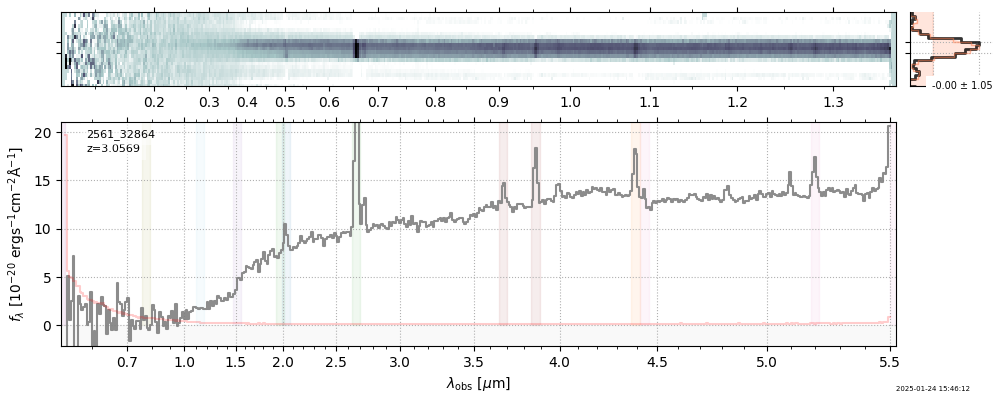
| 30 | uncover-61-v4 | uncover-61-v4_prism-clear_2561_32864.spec.fits | 3.05744 | 11.1156 | 148.446 | 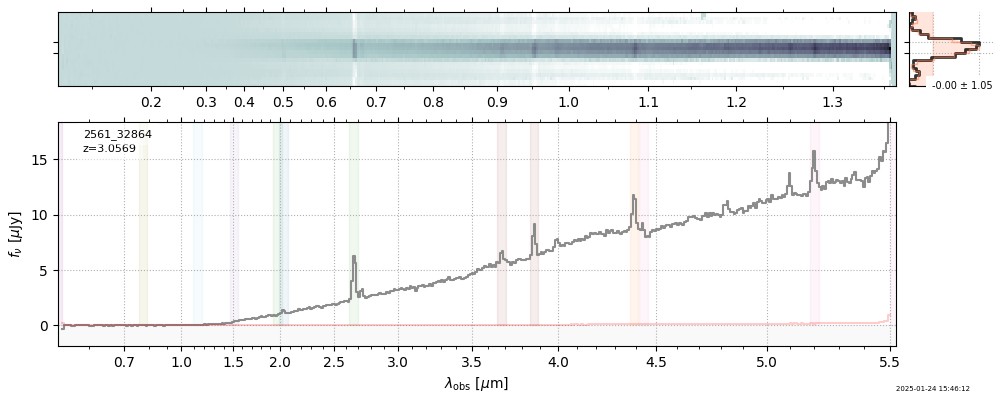 |
 |
||
| 31 | jades-gds-wide3-v4 | jades-gds-wide3-v4_prism-clear_1180_209777.spec.fits | 3.70992 | 11.1048 | 244.425 | 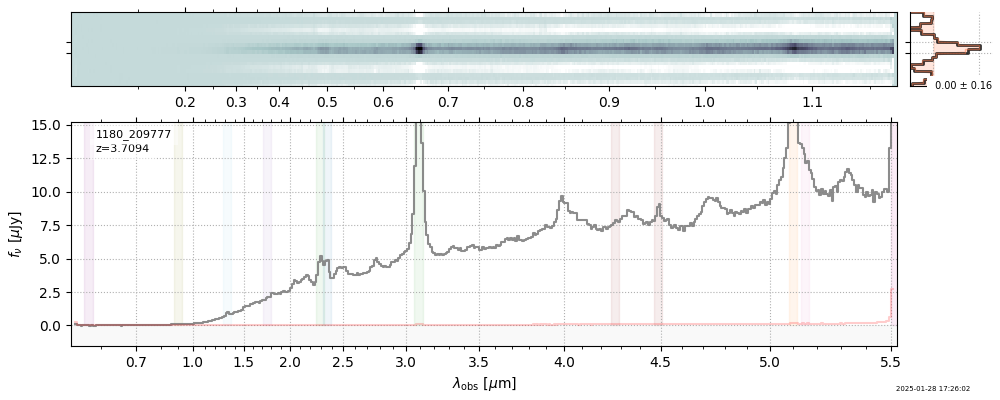 |
 |
Read a spectrum
The spectra can be accessed based on the root and file columns in the summary table.

import msaexp.spectrum
# Set spec_file here, will be used below in NN demo
spec_file = 'rubies-egs61-v4_prism-clear_4233_75646.spec.fits'
row = tab[tab['file'] == spec_file][0]
spec = msaexp.spectrum.SpectrumSampler(FITS_URL.format(**row))
row['Mass']
10.873998434439217
spec.spec.info
<GTable length=473>
name dtype unit description class n_bad
----------- ------- ---- ----------------------------------------- ------------ -----
wave float64 Column 0
flux float64 Column 0
err float64 Column 0
sky float64 uJy MaskedColumn 5
path_corr float64 MaskedColumn 5
npix float64 Column 0
flux_sum float64 Column 0
profile_sum float64 Column 0
var_sum float64 Column 0
corr float64 Column 0
escale float64 Column 0
full_err float64 uJy Column 0
valid bool Column 0
R float64 Spectral resolution from tabulated curves Column 0
to_flam float64 Column 0
plt.plot(spec['wave'], spec['flux'],
label="{file}\nz={z_best:.3f}".format(**row))
plt.legend()
<matplotlib.legend.Legend at 0x1046e1940>
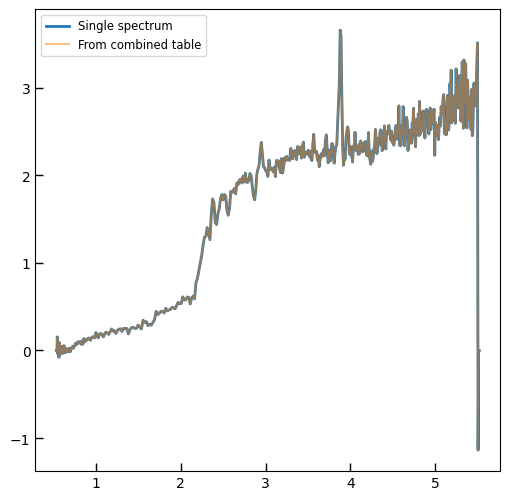
All msaexp PRISM spectra in a single table
All of the 1D extracted spectra have been collated into single FITS tables.
combined_spectra_file = f"dja_msaexp_emission_lines_{version}.prism_spectra.fits"
if os.path.exists(combined_spectra_file):
prism_spectra = utils.read_catalog(combined_spectra_file)
else:
# Combined prism spectra in a single big table (595 Mb)
prism_spectra = utils.read_catalog(
download_file(
f"{URL_PREFIX}/{combined_spectra_file}",
cache=CACHE_DOWNLOADS
),
format='fits',
)
prism_spectra.info()
<GTable length=473>
name dtype shape class n_bad
--------- ------- -------- ------------ ------
wave float64 Column 0
flux float64 (34949,) Column 0
err float64 (34949,) Column 0
sky float64 (34949,) MaskedColumn 697133
path_corr float64 (34949,) MaskedColumn 697133
npix int64 (34949,) Column 0
full_err float64 (34949,) Column 0
valid bool (34949,) Column 0
# The columns of the spectrum have N entries for N objects with a particular grating
# and are aligned with the summary table for that grating
is_prism = tab['grating'] == 'PRISM'
tab['prism_idx'] = 0
tab['prism_idx'][is_prism] = np.arange(is_prism.sum())
print(f"""
File: {combined_spectra_file}\n
{prism_spectra['flux'].shape[0]} wavelength bins\n
PRISM spectra in the merged catalog: {is_prism.sum()}
PRISM spectra in the combined table: {prism_spectra['flux'].shape}
""")
File: dja_msaexp_emission_lines_v4.4.prism_spectra.fits
473 wavelength bins
PRISM spectra in the merged catalog: 34949
PRISM spectra in the combined table: (473, 34949)
# Subset of "valid" spectra defined at most wavelengths
valid_count = prism_spectra['valid'].sum(axis=0)
valid_spec = valid_count > (valid_count.max() - 64)
valid_spec.sum()
31720
row_idx = np.where(tab['file'] == spec_file)[0][0]
row = tab[row_idx]
plt.plot(
spec['wave'], spec['flux'],
lw=2, label='Single spectrum'
)
plt.plot(
prism_spectra['wave'], prism_spectra['flux'][:, row['prism_idx']],
alpha=0.5, label='From combined table'
)
plt.legend()
<matplotlib.legend.Legend at 0x3baa4ea50>
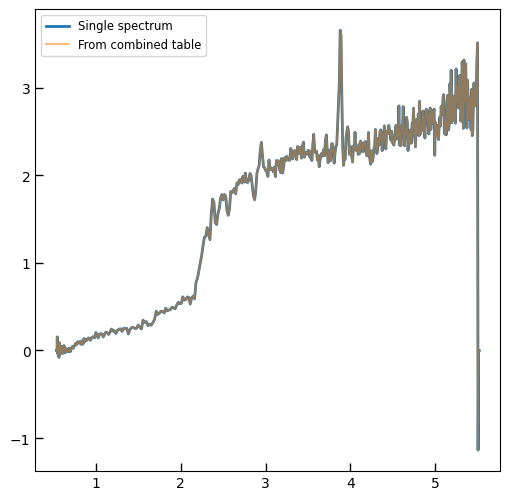
“Stacked” spectrum
Stacking prism spectra isn’t trivial due to the variable dispersion and wavelength sampling. Here just plot a subset on top of each other.
norm_column = 'rest_416_flux'
print(f"Normalization column: '{norm_column}' = {tab[norm_column].description}")
flux_norm = prism_spectra['flux'] / tab[norm_column][is_prism]
# Subset
zi = row['z_best']
dz = 0.05
sample = (tab['z_best'] > zi - dz) & (tab['z_best'] < zi + dz)
sub_sample = sample[is_prism] & valid_spec
sub_idx = np.where(sub_sample)[0]
z_sample = tab['z_best'][is_prism][sample[is_prism] & valid_spec]
fig, axes = plt.subplots(2,1,figsize=(10,7), sharex=False, sharey=True)
for j, z in enumerate(z_sample):
axes[0].plot(
prism_spectra['wave'],
flux_norm[:, sub_idx[j]],
alpha=0.1
)
axes[1].plot(
prism_spectra['wave'] / (1 + z),
flux_norm[:, sub_idx[j]],
alpha=0.1
)
axes[0].set_ylim(-1, 10)
Normalization column: 'rest_416_flux' = Spectrum flux in synthetic_i
(-1.0, 10.0)
“Nearest neighbor” spectra
Simple “nearest neighbors” of the observed-frame normalized spectra extracted from a KDTree.
fix_flux_norm = flux_norm*1.
fix_flux_norm[~np.isfinite(flux_norm)] = 0
tr = cKDTree(fix_flux_norm[:, valid_spec].T)
N_nn = 32
row = tab[tab['file'] == spec_file][0]
# Features matrix is full normalized observed-frame spectra
Xfeatures = fix_flux_norm
tr_ds, tr_idx = tr.query(Xfeatures[:, row['prism_idx']], k=N_nn)
display_columns = [
'root','file','zrf','Mass','ha_eqw_with_limits','Thumb','Slit_Thumb','Spectrum_fnu', 'Spectrum_flam'
]
df = tab[display_columns][is_prism][valid_spec][tr_idx][:16].to_pandas()
display(Markdown(df.to_markdown()))
| root | file | zrf | Mass | ha_eqw_with_limits | Thumb | Slit_Thumb | Spectrum_fnu | Spectrum_flam | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | rubies-egs61-v4 | rubies-egs61-v4_prism-clear_4233_75646.spec.fits | 4.90238 | 10.874 | 24.3336 |  |
 |
||
| 1 | gds-barrufet-s67-v4 | gds-barrufet-s67-v4_prism-clear_2198_8777.spec.fits | 4.65301 | 10.6769 | 8.26113 |  |
 |
||
| 2 | gds-barrufet-s67-v4 | gds-barrufet-s67-v4_prism-clear_2198_8290.spec.fits | 4.34034 | 10.5556 | 4.0581 |  |
 |
||
| 3 | mom-uds02-v4 | mom-uds02-v4_prism-clear_5224_144670.spec.fits | 3.97196 | 10.6169 | 14.667 | 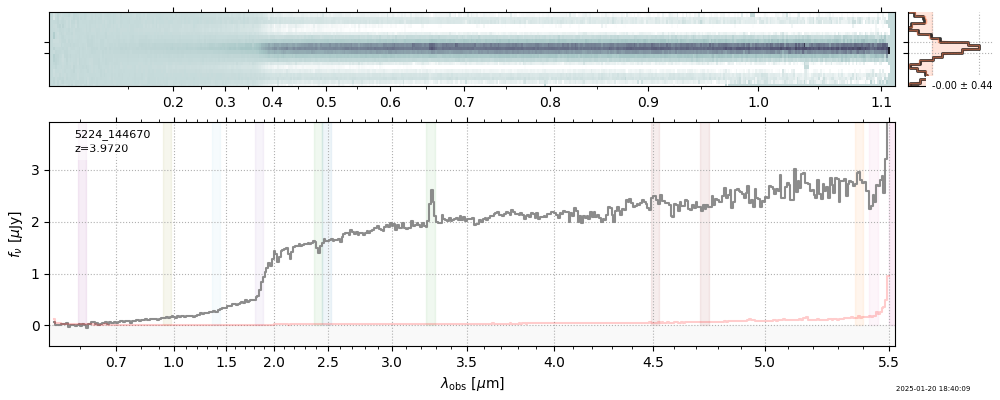 |
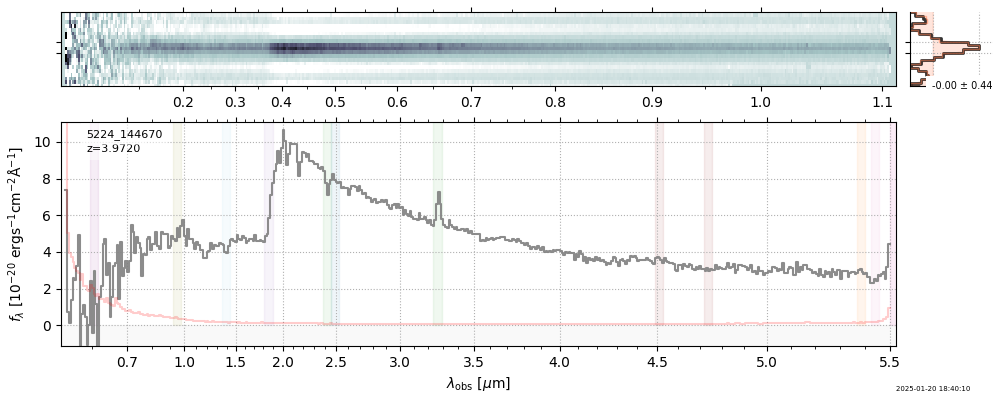 |
||
| 4 | jades-gdn09-v4 | jades-gdn09-v4_prism-clear_1181_72127.spec.fits | 4.13556 | 10.5771 | 75.7974 | 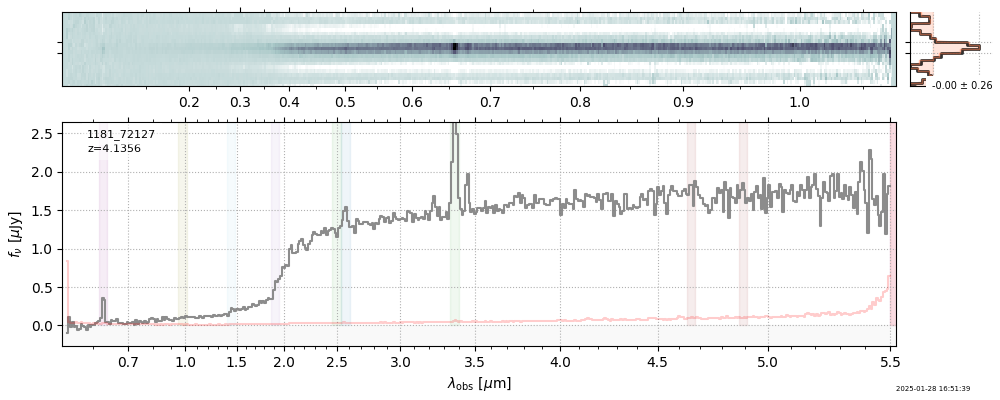 |
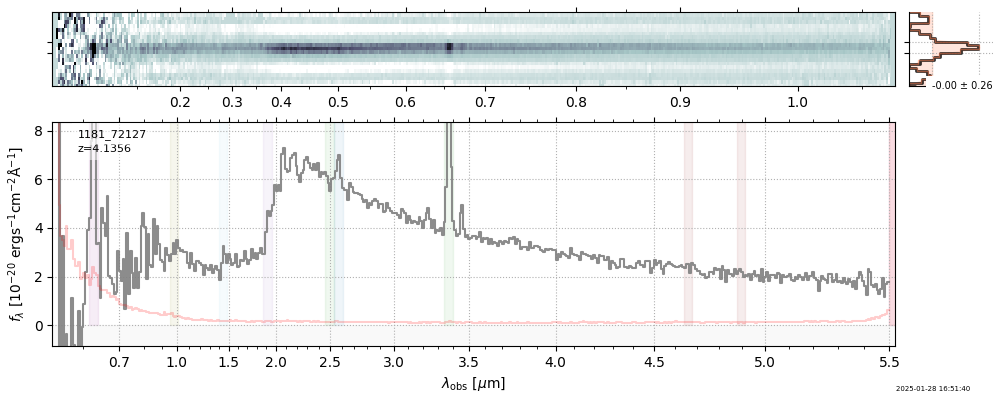 |
||
| 5 | glazebrook-cos-obs1-v4 | glazebrook-cos-obs1-v4_prism-clear_2565_10559.spec.fits | 4.28971 | 10.5518 | 3.80496 |  |
 |
||
| 6 | jades-gds-wide-v4 | jades-gds-wide-v4_prism-clear_1180_12619.spec.fits | 3.60465 | 10.6324 | 27.8163 | 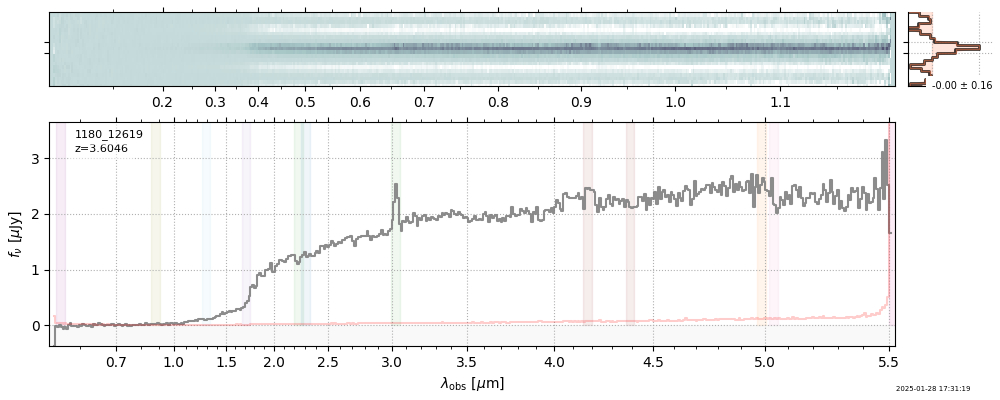 |
 |
||
| 7 | rubies-uds2-v4 | rubies-uds2-v4_prism-clear_b28.spec.fits | 4.39429 | 10.8802 | 5.61715 |  |
 |
||
| 8 | rubies-uds2-v4 | rubies-uds2-v4_prism-clear_4233_b28.spec.fits | 4.39461 | 10.8801 | 5.59215 |  |
 |
||
| 9 | glazebrook-cos-obs3-v4 | glazebrook-cos-obs3-v4_prism-clear_2565_20115.spec.fits | 3.72605 | 11.2231 | 2.61723 |  |
 |
||
| 10 | gto-wide-egs1-v4 | gto-wide-egs1-v4_prism-clear_1213_4358.spec.fits | 4.29302 | 10.6918 | 21.9825 |  |
 |
||
| 11 | jades-gdn09-v4 | jades-gdn09-v4_prism-clear_1181_80660.spec.fits | 4.40673 | 10.2177 | 25.751 | 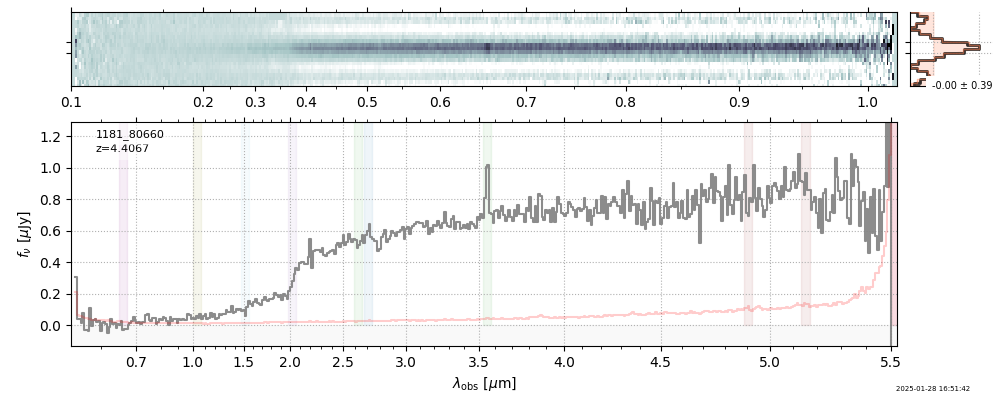 |
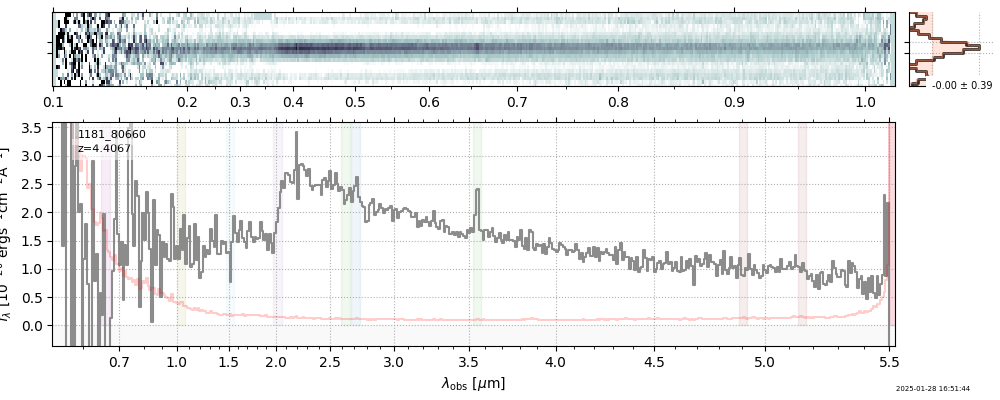 |
||
| 12 | glazebrook-v4 | glazebrook-v4_prism-clear_2565_10459.spec.fits | 3.97084 | 10.6829 | 4.65304 |  |
 |
||
| 13 | jades-gdn198-v4 | jades-gdn198-v4_prism-clear_1181_76320.spec.fits | 3.24558 | 10.306 | 5.3213 | 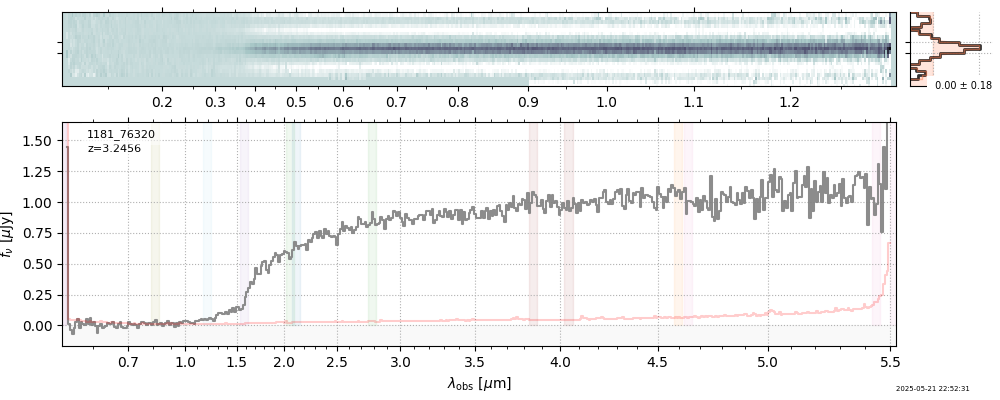 |
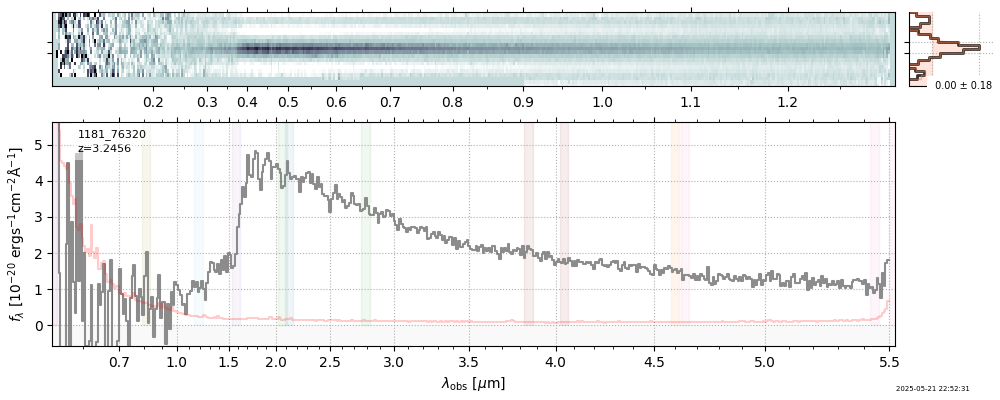 |
||
| 14 | macs1149-v4 | macs1149-v4_prism-clear_1208_5103925.spec.fits | 3.69631 | 10.8235 | 6.45299 | 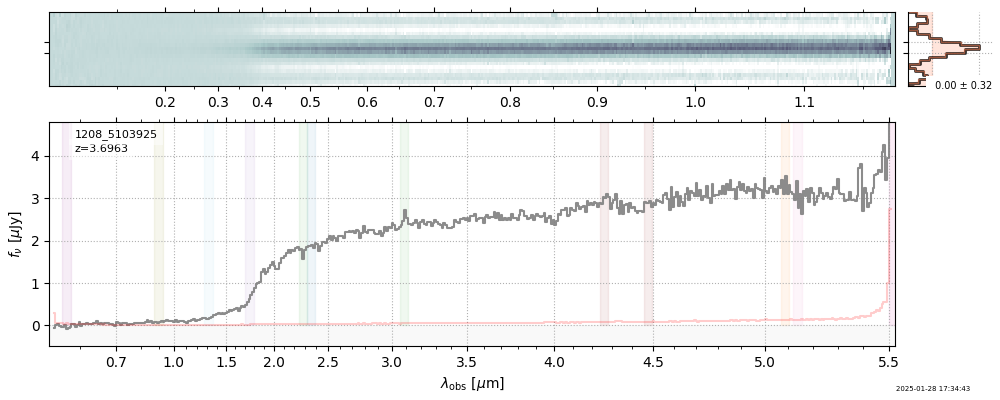 |
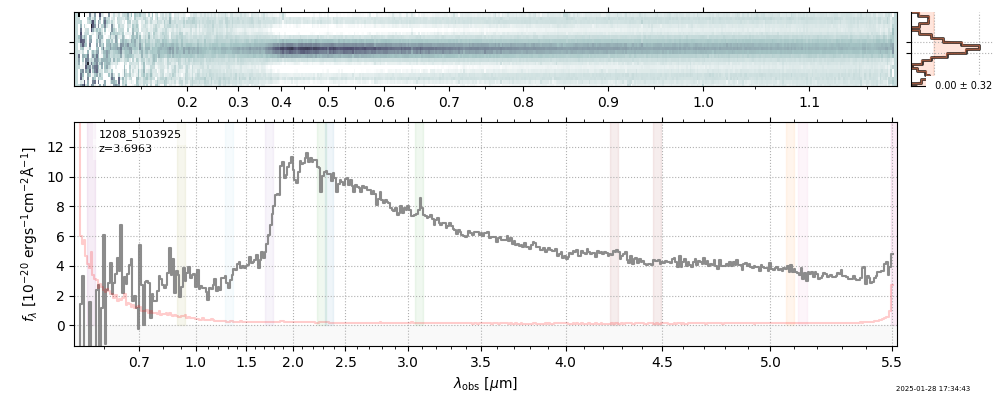 |
||
| 15 | jades-gdn-v4 | jades-gdn-v4_prism-clear_1181_76320.spec.fits | 3.23434 | 10.3316 | 4.69578 | 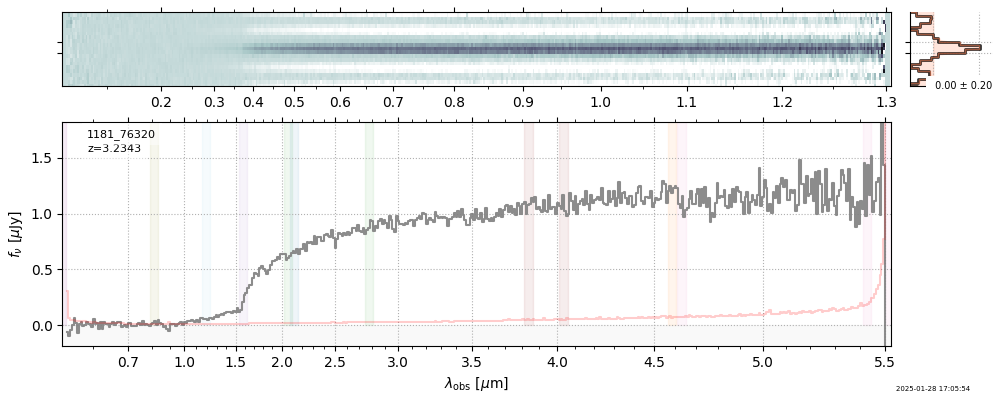 |
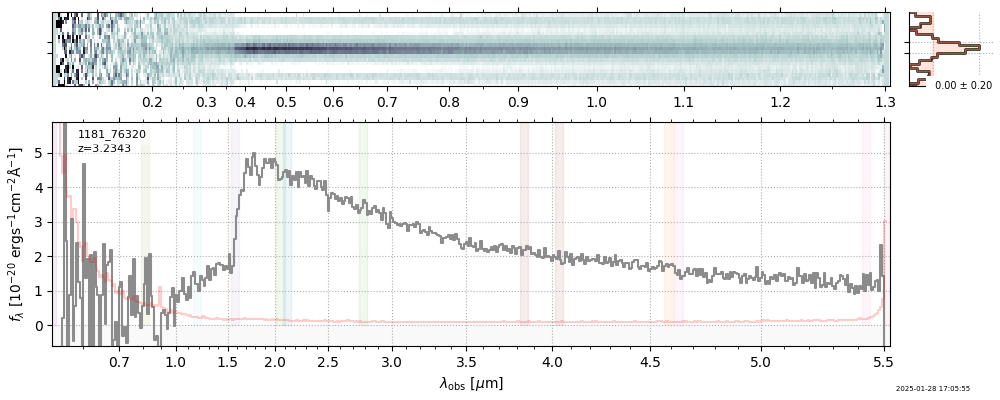 |
Compute NN in rest-frame
The example above computed “raw” nearest neighbors from the full observed-frame spectra. To compute NN in the rest-frame, interpolate spectra to a fixed rest-frame wavelength grid.
from tqdm import tqdm
# rest-frame interpolated
wrest = utils.log_zgrid([0.08, 1.4], 1./1200/np.log(10))
wrest = np.unique([prism_spectra['wave'] / (1+z) for z in utils.log_zgrid([0.5, 7], 0.3)])
print(wrest.shape)
zero = wrest*0.
rest_flux_norm = []
for i, z in tqdm(enumerate(tab['zrf'][is_prism])):
if z < 0:
rest_flux_norm.append(zero)
else:
rest_flux_norm.append(np.interp(wrest, prism_spectra['wave'] / (1 + z), flux_norm[:,i], left=0, right=0))
rest_flux_norm = np.array(rest_flux_norm).T
rest_flux_norm[~np.isfinite(rest_flux_norm)] = 0
(2838,)
34949it [00:02, 14248.53it/s]
fig, ax = plt.subplots(1,1,figsize=(8,5))
ax.plot(
prism_spectra['wave'] / (1 + tab['zrf'][row_idx]), flux_norm[:, row['prism_idx']],
alpha=0.5, label='From combined table',
color='0.5',
)
valid_rest = rest_flux_norm[:, row['prism_idx']] != 0
ax.scatter(
wrest[valid_rest],
rest_flux_norm[:, row['prism_idx']][valid_rest],
marker='.',
color='purple',
label='resampled rest-frame',
alpha=0.5
)
ax.legend()
fig.tight_layout(pad=1)
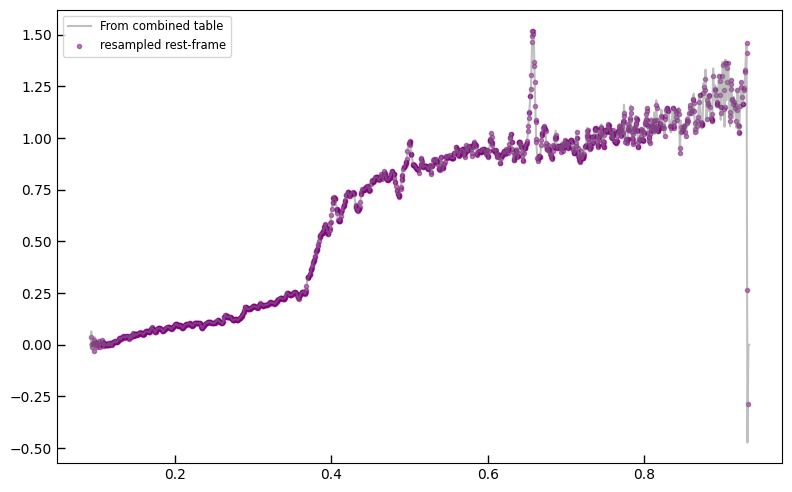
# Compute nearest-neighbors over limited rest-frame wavelength range
sli = np.where((wrest > 0.2) & (wrest < 0.8))[0]
Xfeatures = rest_flux_norm[sli,:]
tr = cKDTree(Xfeatures[:, valid_spec].T)
if False:
# Trimmed spectrum and include log(1+z) as a feature
sli = slice(32, -16)
fix_flux_norm = np.vstack([flux_norm[sli,:]**1, np.log(1+tab['zrf'][is_prism]) * 1e-2])
fix_flux_norm[~np.isfinite(fix_flux_norm)] = 0
Xfeatures = fix_flux_norm
tr = cKDTree(Xfeatures[:, valid_spec].T)
row = tab[tab['file'] == spec_file][0]
tr_ds, tr_idx = tr.query(Xfeatures[:, row['prism_idx']], k=N_nn)
df = tab[display_columns][is_prism][valid_spec][tr_idx][:16].to_pandas()
display(Markdown(df.to_markdown()))
| root | file | zrf | Mass | ha_eqw_with_limits | Thumb | Slit_Thumb | Spectrum_fnu | Spectrum_flam | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | rubies-egs61-v4 | rubies-egs61-v4_prism-clear_4233_75646.spec.fits | 4.90238 | 10.874 | 24.3336 |  |
 |
||
| 1 | ceers-v4 | ceers-v4_prism-clear_1345_2759.spec.fits | 3.43971 | 10.622 | 31.5176 | 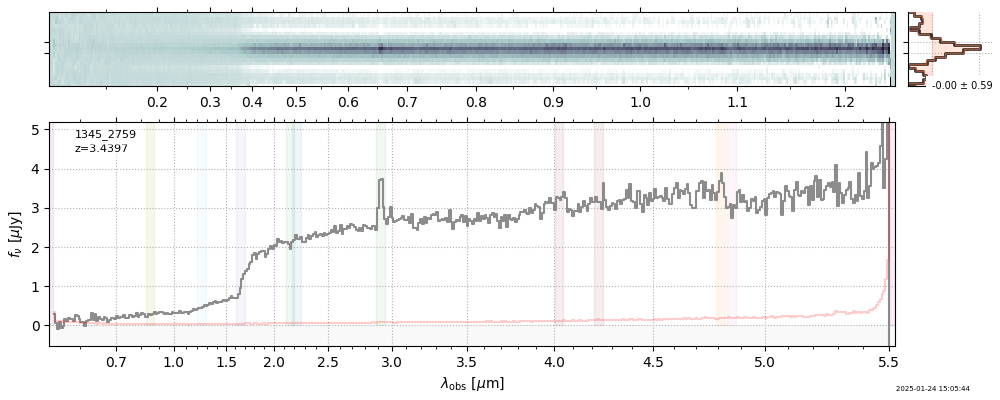 |
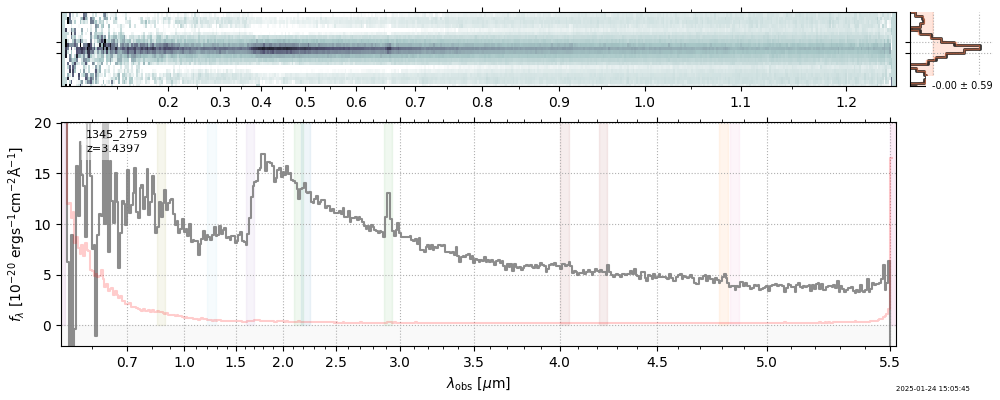 |
||
| 2 | capers-cos16-v4 | capers-cos16-v4_prism-clear_6368_24202.spec.fits | 3.09955 | 10.9213 | 20.8004 | 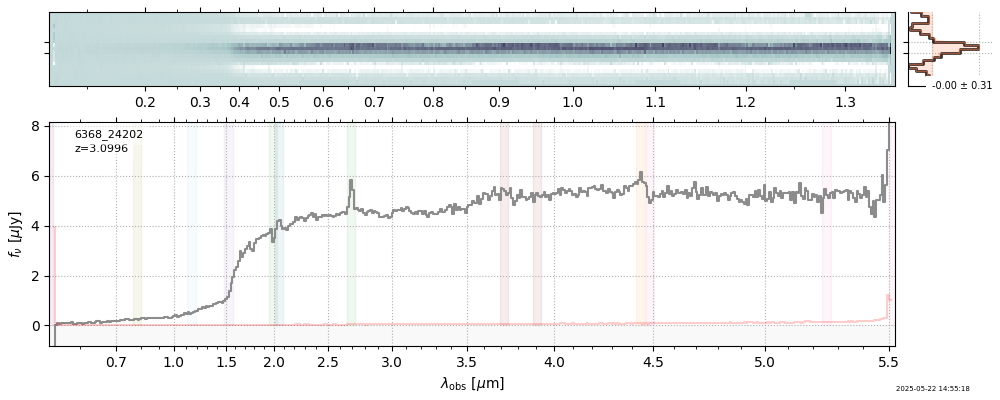 |
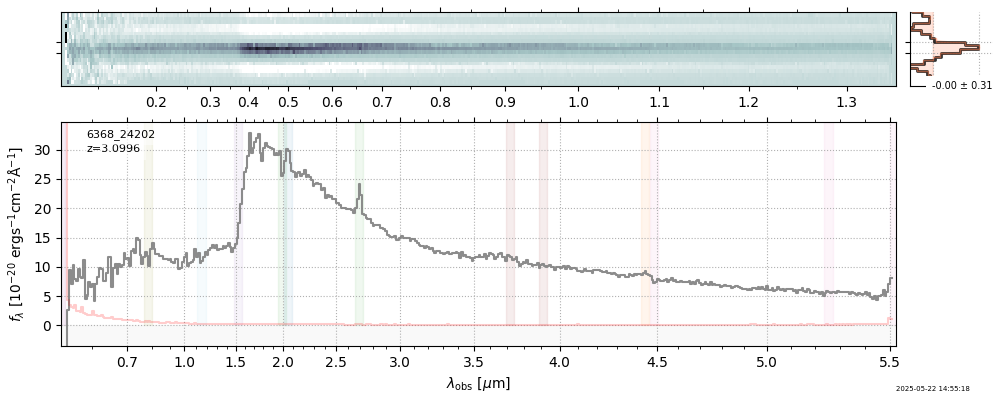 |
||
| 3 | mom-uds02-v4 | mom-uds02-v4_prism-clear_5224_144670.spec.fits | 3.97196 | 10.6169 | 14.667 |  |
 |
||
| 4 | nexus-obs5-v4 | nexus-obs5-v4_prism-clear_5105_27813.spec.fits | 3.96582 | 11.0988 | 8.80705 | 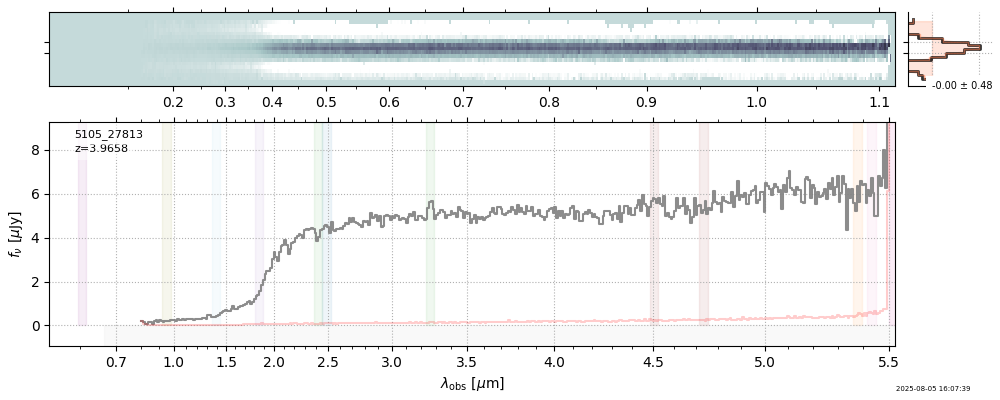 |
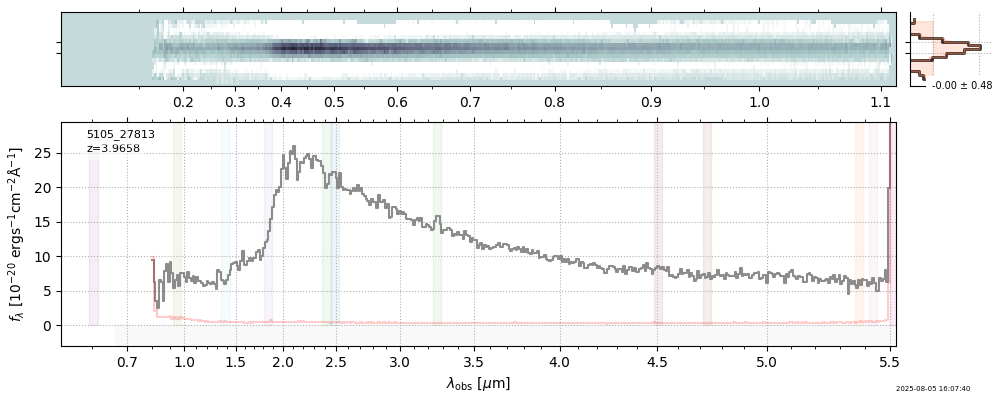 |
||
| 5 | jades-gds05-v4 | jades-gds05-v4_prism-clear_1286_194373.spec.fits | 2.67694 | 10.2399 | 42.391 |  |
 |
||
| 6 | ceers-ddt-v4 | ceers-ddt-v4_prism-clear_2750_307.spec.fits | 2.93255 | 10.9374 | 2.72842 | 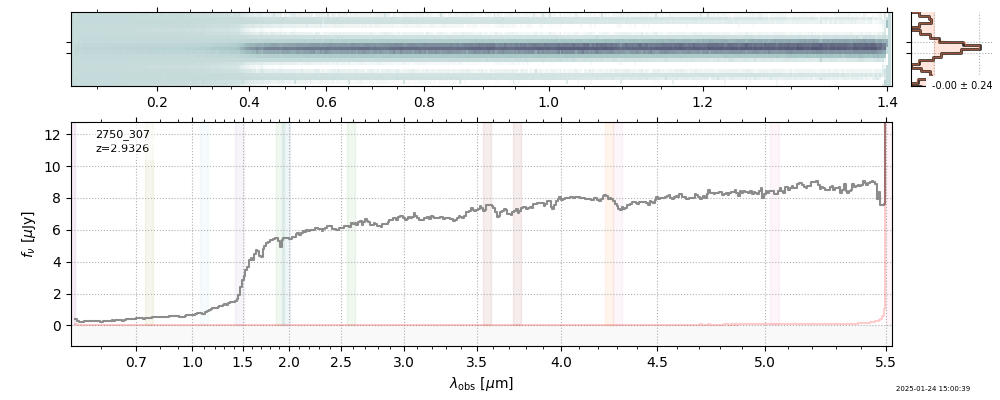 |
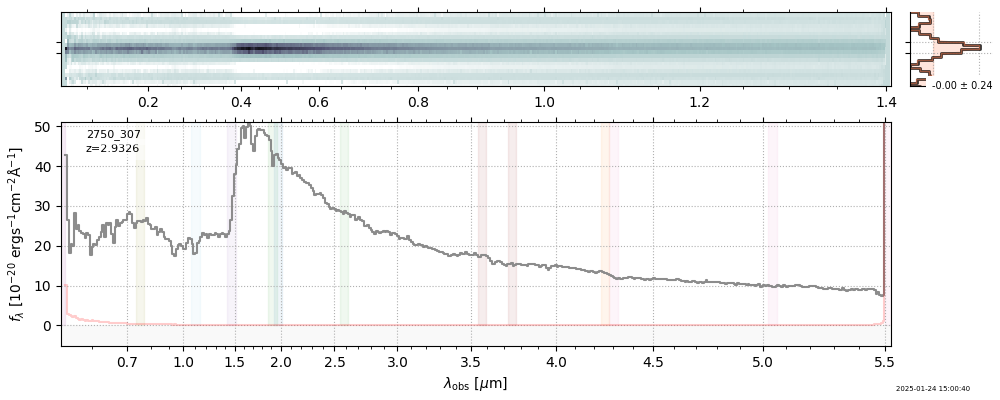 |
||
| 7 | glazebrook-egs-v4 | glazebrook-egs-v4_prism-clear_2565_18996.spec.fits | 3.23542 | 10.9906 | 23.3237 | 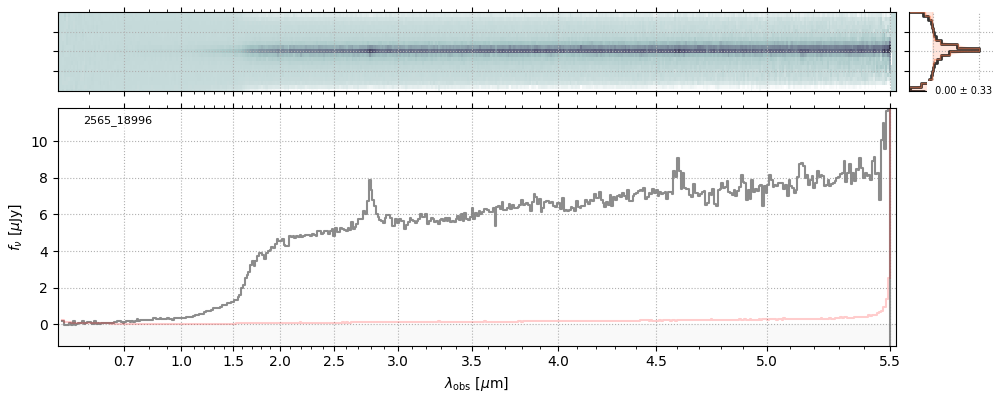 |
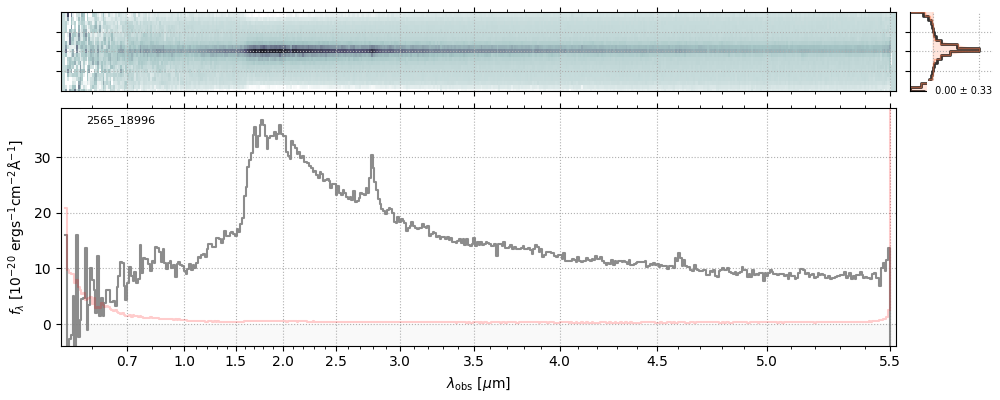 |
||
| 8 | capers-egs49-v4 | capers-egs49-v4_prism-clear_6368_7806.spec.fits | 3.45255 | 10.285 | 33.2528 |  |
 |
||
| 9 | ceers-v4 | ceers-v4_prism-clear_1345_2779.spec.fits | 3.23845 | 10.8818 | 31.1324 | 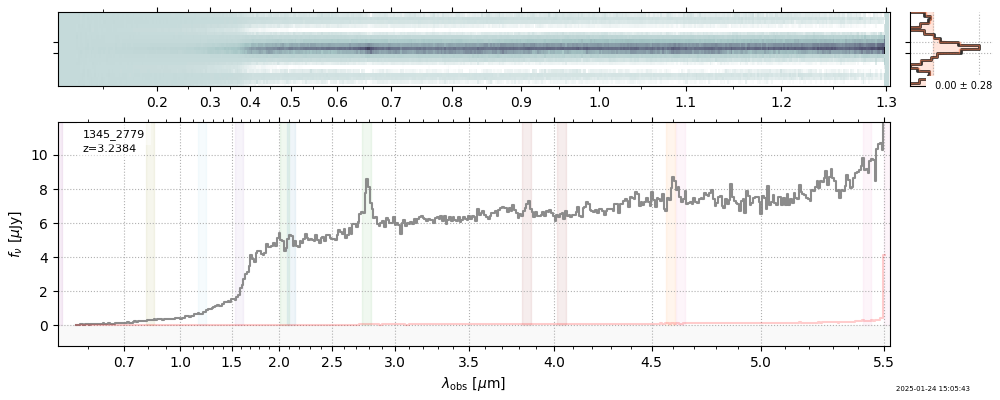 |
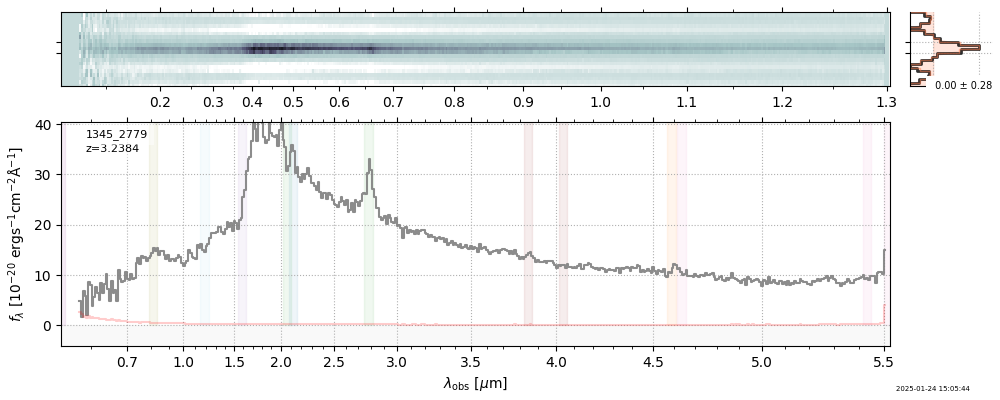 |
||
| 10 | snh0pe-v4 | snh0pe-v4_prism-clear_4446_274.spec.fits | 4.11111 | 10.8132 | 4.80224 |  |
 |
||
| 11 | egs-nelsonx-v4 | egs-nelsonx-v4_prism-clear_4106_76085.spec.fits | 3.22192 | 10.5733 | 34.906 | 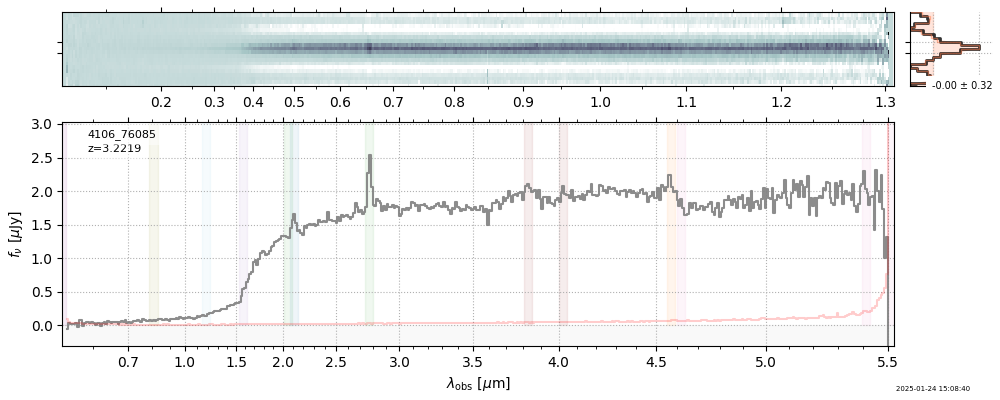 |
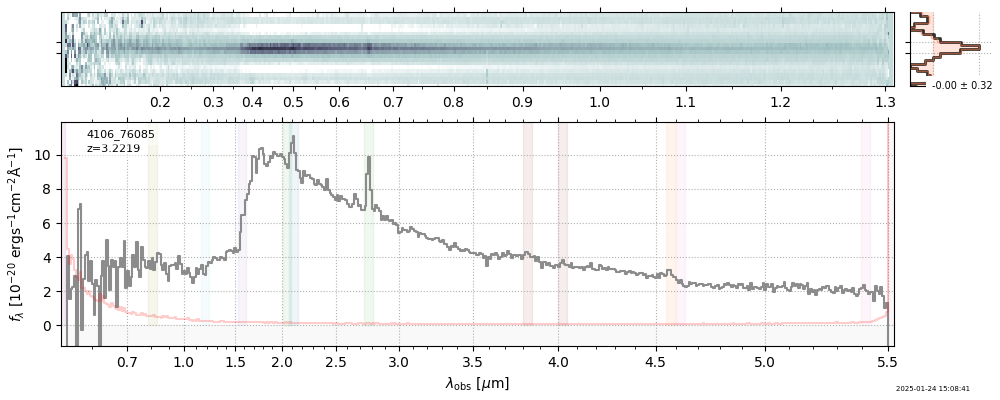 |
||
| 12 | rubies-egs63-v4 | rubies-egs63-v4_prism-clear_4233_58841.spec.fits | 3.45082 | 10.4517 | 27.5816 | 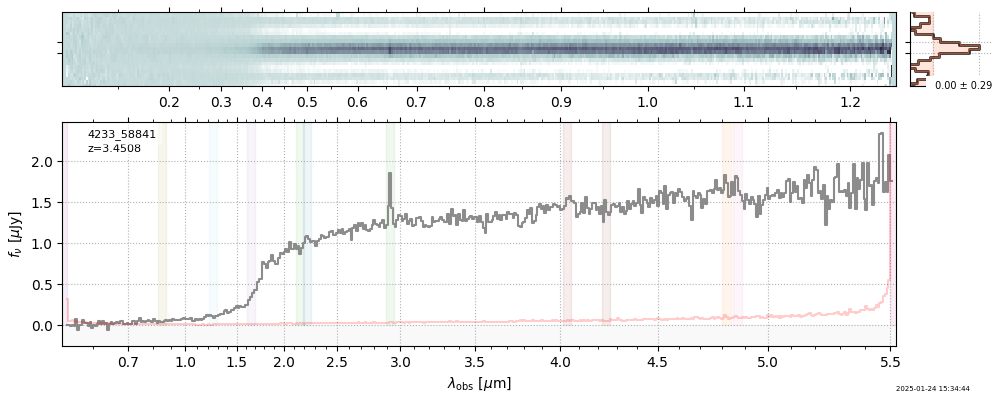 |
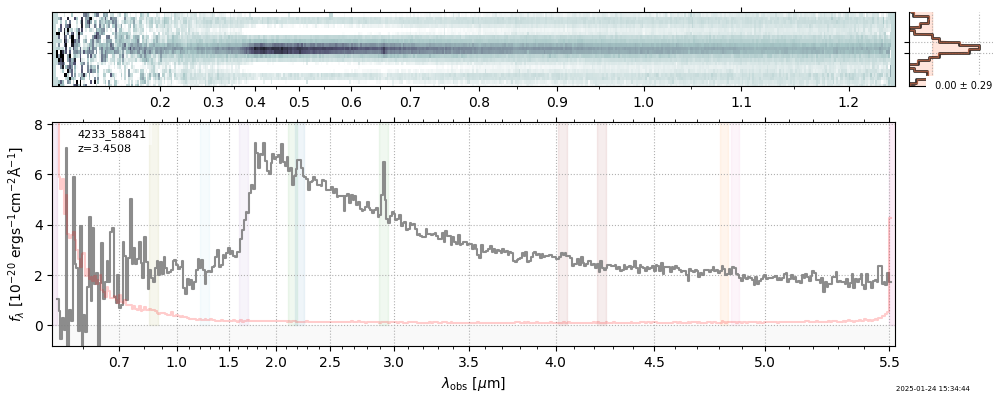 |
||
| 13 | rubies-egs63-v4 | rubies-egs63-v4_prism-clear_4233_61168.spec.fits | 3.43539 | 10.8289 | 3.56196 | 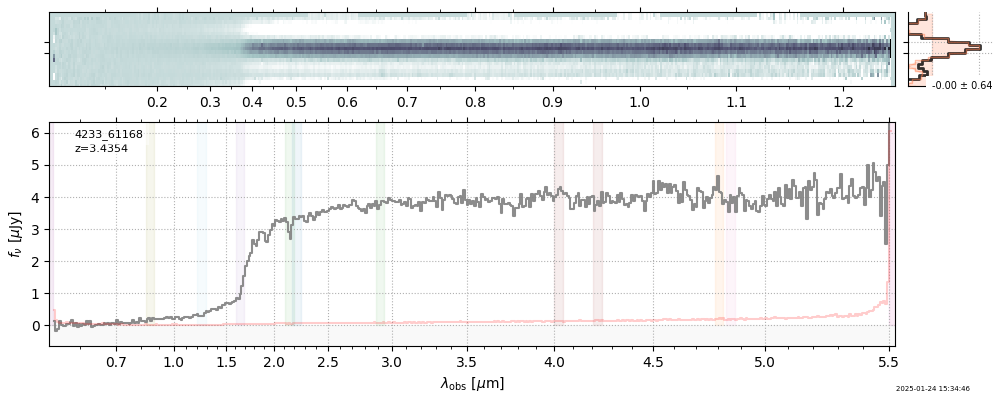 |
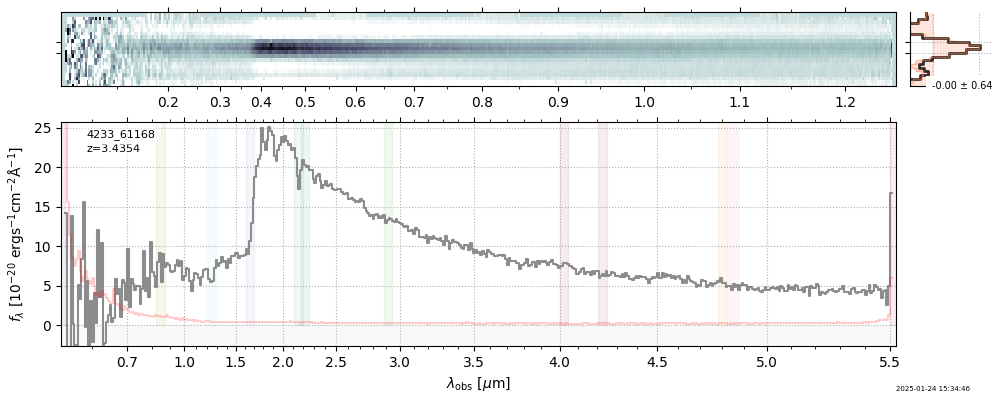 |
||
| 14 | glazebrook-egs-v4 | glazebrook-egs-v4_prism-clear_2565_31322.spec.fits | 3.4242 | 10.961 | 3.01521 | 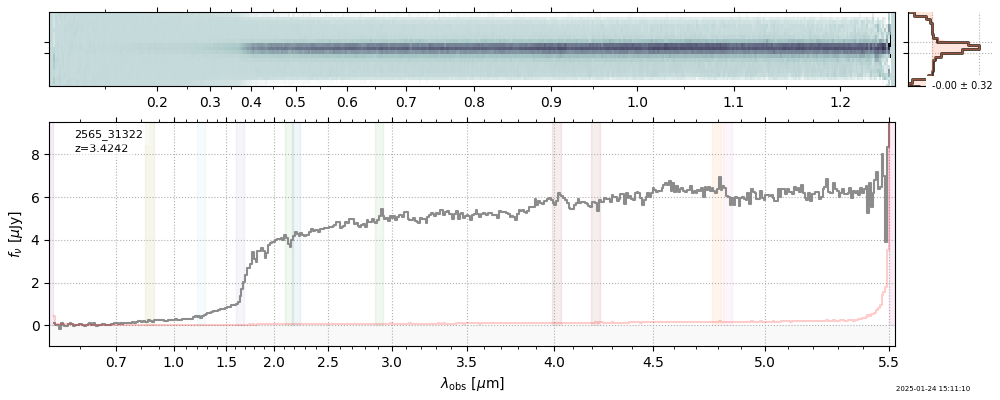 |
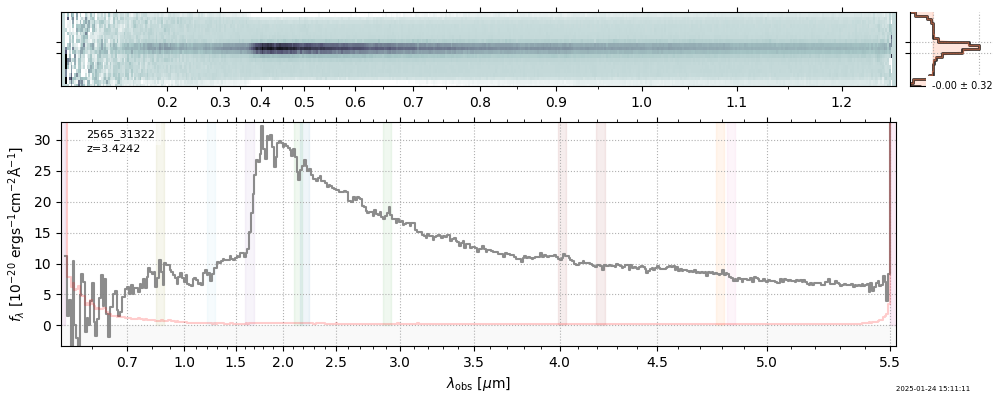 |
||
| 15 | jades-gds03-v4 | jades-gds03-v4_prism-clear_1286_10026167.spec.fits | 3.50452 | 10.3615 | 2.95476 | 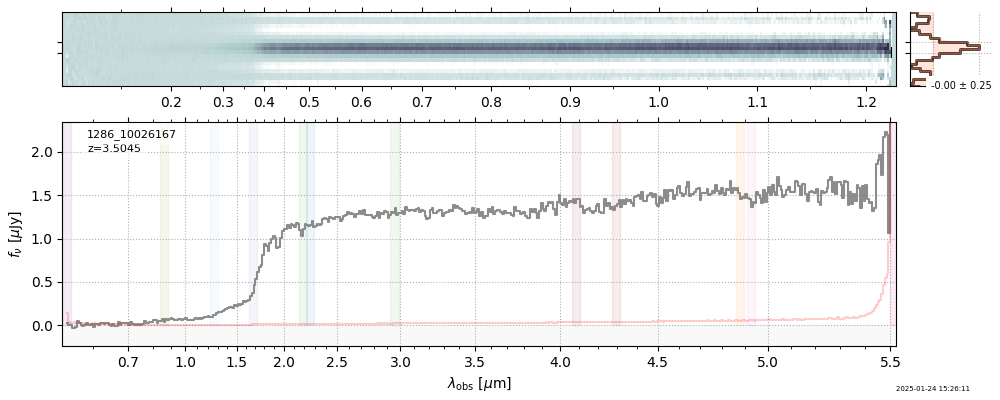 |
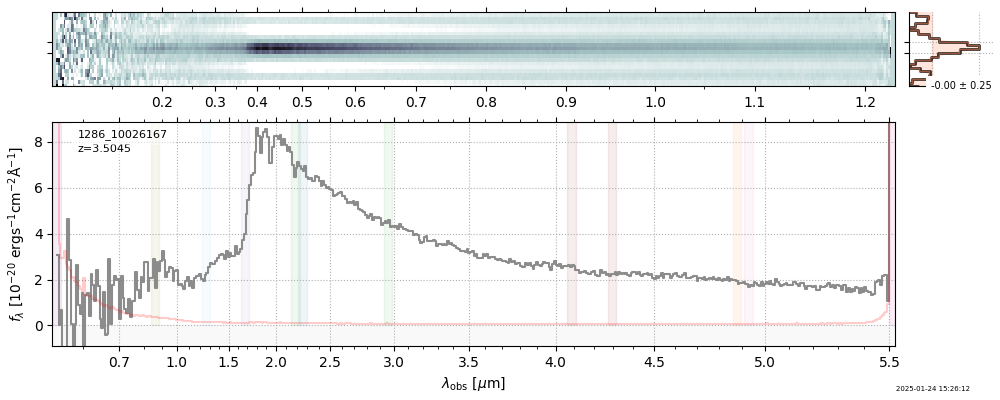 |
nn_sample = tab['ra'] < 0
nn_j = np.where(is_prism)[0][valid_spec][tr_idx]
nn_sample[nn_j] = True
print(nn_sample.sum())
sub_sample = nn_sample[is_prism] & valid_spec
sub_idx = np.where(sub_sample)[0]
z_sample = tab['zrf'][is_prism][nn_sample[is_prism] & valid_spec]
file_sample = tab['file'][is_prism][nn_sample[is_prism] & valid_spec]
fig, axes = plt.subplots(2,1,figsize=(10,7), sharex=False, sharey=True)
flam = -2
if 0:
flam = 0
for j, z in enumerate(z_sample):
kws = dict(
alpha=0.3 if file_sample[j] == spec_file else 0.1,
color='k' if file_sample[j] == spec_file else plt.cm.plasma(j/len(z_sample)),
label=spec_file if file_sample[j] == spec_file else None,
zorder=1000 if file_sample[j] == spec_file else 10,
)
axes[0].plot(
# prism_spectra['wave'],
# (flux_norm[:, sub_idx[j]] * (prism_spectra['wave'] / (1 + z) / 0.7)**flam),
(Xfeatures[:, sub_idx[j]]), # * (prism_spectra['wave'] / (1 + z) / 0.7)**flam),
**kws
)
axes[1].plot(
prism_spectra['wave'] / (1 + z),
(flux_norm[:, sub_idx[j]] * (prism_spectra['wave'] / (1 + z) / 0.7)**flam),
**kws
)
axes[0].set_xlabel(r'$i$')
axes[0].set_ylabel(r'$X_i$')
ymax = 2.2
axes[1].legend(loc='upper right')
axes[1].set_xlabel(r'$\lambda_\mathrm{rest}$')
axes[1].set_ylabel('normalized spectrum')
axes[0].set_ylim(-0.1*ymax, ymax)
for ax in axes:
ax.grid()
fig.tight_layout(pad=1)
32
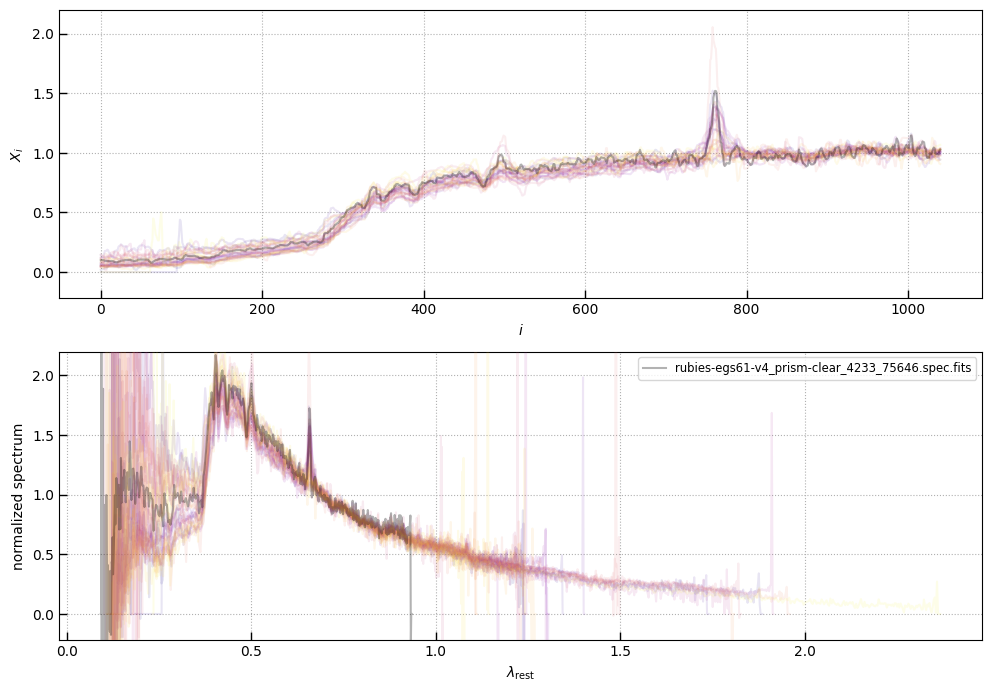
fig, ax = plt.subplots(1,1,figsize=(9,6))
tr_spec = (rest_flux_norm[:, valid_spec][:,tr_idx].T * (wrest/0.7)**flam).T
tr_spec[np.abs(tr_spec) < 1.e-6] = np.nan
tr_spec = tr_spec[:,:16]
xpl = wrest
xpl = np.arange(len(xpl))
_ = ax.plot(xpl, tr_spec[:,1:], alpha=0.1, color='0.7')
# _ = ax.plot(wrest, np.nanmean(tr_spec, axis=1), alpha=0.5, color='k')
_ = ax.plot(xpl, np.nanmedian(tr_spec[:,1:], axis=1), alpha=0.5, color='k', label='NN median')
src_spec = tr_spec[:,0] #np.nanmean((rest_flux_norm[:, valid_spec][sli,:][:,tr_idx[:1]].T * (wrest[sli]/0.7)**flam).T, axis=1)
_ = ax.plot(xpl, src_spec, alpha=0.6, color='tomato', label=spec_file)
ymax = 1.5*np.nanpercentile(src_spec, 90)
xt = np.append(np.arange(0.1, 1.01, 0.1), np.arange(1.2, 1.81, 0.2))
ax.set_xticks(np.interp(xt, wrest, xpl))
ax.set_xticklabels([f'{v:.1f}' for v in xt])
# ax.set_xlim(*np.interp([0.3, 1.9], wrest, xpl))
ax.legend(loc='upper right')
ax.set_ylim(-0.1*ymax, ymax)
ax.grid()
# ax.semilogx()
ax.set_ylabel(r'$f_\lambda$')
ax.set_xlabel(r'$\lambda_\mathrm{rest}$')
fig.tight_layout(pad=1)
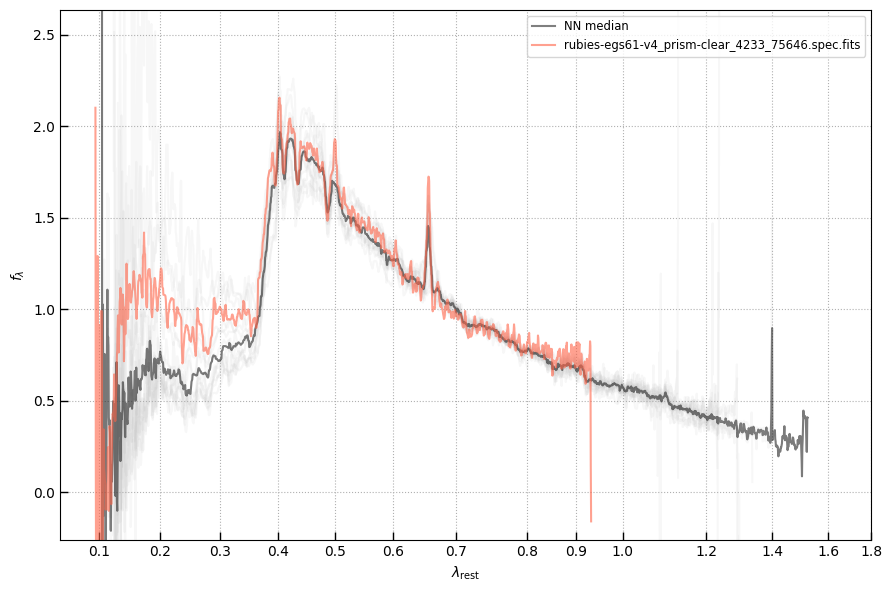
Merged 1D grating spectra
Read 1D grating arrays
try:
_ = grating_spectra
except NameError:
# Initialize
grating_spectra = {}
for (grating, filter) in [('G140M', 'F070LP'), ('G235M','F170LP'), ('G395M', 'F290LP')]:
key = (grating, filter)
grating_spectra_file = f"dja_msaexp_emission_lines_{version}.{grating}-{filter}_spectra.fits".lower()
if key not in grating_spectra:
print(f"{key}: load {grating_spectra_file}")
if os.path.exists(grating_spectra_file):
grating_spectra[key] = utils.read_catalog(grating_spectra_file)
else:
# Combined grating spectra in a single big table
grating_spectra[key] = utils.read_catalog(
download_file(
f"{URL_PREFIX}/{grating_spectra_file}",
cache=CACHE_DOWNLOADS
),
format='fits',
)
else:
print(f"{key} spectra already loaded from {grating_spectra_file}")
is_grating = (tab["grating"] == grating) & (tab["filter"] == filter)
print(f"""
{grating_spectra[key]["flux"].shape[0]} wavelength bins\n
{grating} {filter} spectra in the combined table: {grating_spectra[key]["flux"].shape}
{grating} {filter} entries in the summary table: {is_grating.sum()}
""")
('G140M', 'F070LP'): load dja_msaexp_emission_lines_v4.4.g140m-f070lp_spectra.fits
4667 wavelength bins
G140M F070LP spectra in the combined table: (4667, 5851)
G140M F070LP entries in the summary table: 5851
('G235M', 'F170LP'): load dja_msaexp_emission_lines_v4.4.g235m-f170lp_spectra.fits
3685 wavelength bins
G235M F170LP spectra in the combined table: (3685, 8000)
G235M F170LP entries in the summary table: 8000
('G395M', 'F290LP'): load dja_msaexp_emission_lines_v4.4.g395m-f290lp_spectra.fits
1661 wavelength bins
G395M F290LP spectra in the combined table: (1661, 13606)
G395M F290LP entries in the summary table: 13606
NN with grating spectra
Look for grating spectra of sources identified as nearest-neighbors above, matching on the obsid unique identifier.
nn_objid = tab['objid'][is_prism][valid_spec][tr_idx]
# gratings = np.unique(tab['grating'])
match_objid = np.isin(tab['objid'], nn_objid) & (tab['line_ha_err'] > 0)
_ = utils.Unique(tab['grating'][match_objid])
_ = utils.Unique(tab['filter'][match_objid])
N value
==== ==========
2 G140M
2 G235H
8 G235M
3 G395H
10 G395M
N value
==== ==========
2 F070LP
10 F170LP
13 F290LP
from scipy.stats import binned_statistic
import msaexp.utils
fig, axes = plt.subplots(1,3,figsize=(10,5), width_ratios=[0.6, 0.3, 1], sharey=True)
weighted_mean = {}
for key in grating_spectra:
grating, filter = key
if grating == 'G140M':
continue
full_spec = []
in_grating = (tab['grating'] == grating) & (tab['filter'] == filter)
# Sample with grating spectra that cover H-alpha
nn_with_grating = np.isin(tab['objid'][in_grating], nn_objid)
nn_with_grating &= (tab['line_ha_err'][in_grating] > 0) | (tab['line_oiii_5007_err'][in_grating] > 0)
if nn_with_grating.sum() == 0:
continue
print(f'{grating}-{filter} N={nn_with_grating.sum()}')
for j in np.where(nn_with_grating)[0]:
wrest_j = grating_spectra[grating, filter]['wave'] / (1 + tab['z_best'][in_grating][j])
flux_j = grating_spectra[grating, filter]['flux'][:,j] * 1
err_j = grating_spectra[grating, filter]['err'][:,j] * 1
wsub = (wrest_j > 0.64) & (wrest_j < 0.68) & (err_j > 0)
renorm_flux = np.median(flux_j[wsub])
if 0:
# Normalize to prism i band
k = is_prism & (tab['objid'] == tab['objid'][in_grating][j])
renorm_flux = np.nanmean(tab['rest_415_flux'][k])
# print(renorm_flux, wsub.sum())
# renorm_flux = 1.0
flux_j /= renorm_flux
err_j /= renorm_flux
flux_j[err_j <= 0] = np.nan
err_j[err_j <= 0] = np.nan
# if np.nanmedian(err_j[wsub]) > 0.5:
# # Skip low S/N
# continue
full_spec.append([wrest_j, flux_j, err_j])
for ax in axes:
ax.step(
wrest_j,
flux_j,
alpha=0.02,
zorder=-1,
)
# Bin by grating
full_spec = np.array(full_spec)
target_R = 1500
wbin = 10**np.arange(*np.log10([0.3, 1.0]), 1./target_R/np.log(10))
wbin_edge = msaexp.utils.array_to_bin_edges(wbin)
wht = 1. / (full_spec[:,2,:]**2 + (0.1*full_spec[:,1,:])**2)
num = binned_statistic(
full_spec[:,0,:].flatten(),
(full_spec[:,1,:] * wht).flatten(),
bins=wbin_edge,
statistic=np.nansum
)
denom = binned_statistic(
full_spec[:,0,:].flatten(),
wht.flatten(),
bins=wbin_edge,
statistic=np.nansum
)
wflux = num.statistic / denom.statistic
werr = np.sqrt(1./denom.statistic)
weighted_mean[key] = (wbin, wflux, werr)
# optionally trim low S/N
trim = wflux > 5 * werr
for ax in axes:
ax.step(wbin[trim], wflux[trim], color='k', alpha=0.3)
axes[0].set_xlim(0.31, 0.64)
axes[1].set_xlim(0.648, 0.665)
axes[2].set_xlim(0.62, 0.96)
ax.set_ylim(*(np.array([-0.03, 1.02])*2))
axes[0].set_ylabel(r'normalized $f_\nu$')
axes[1].set_xlabel(r'$\lambda_\mathrm{rest}$')
for ax in axes:
ax.grid()
fig.tight_layout(pad=1)
G235M-F170LP N=9
G395M-F290LP N=10
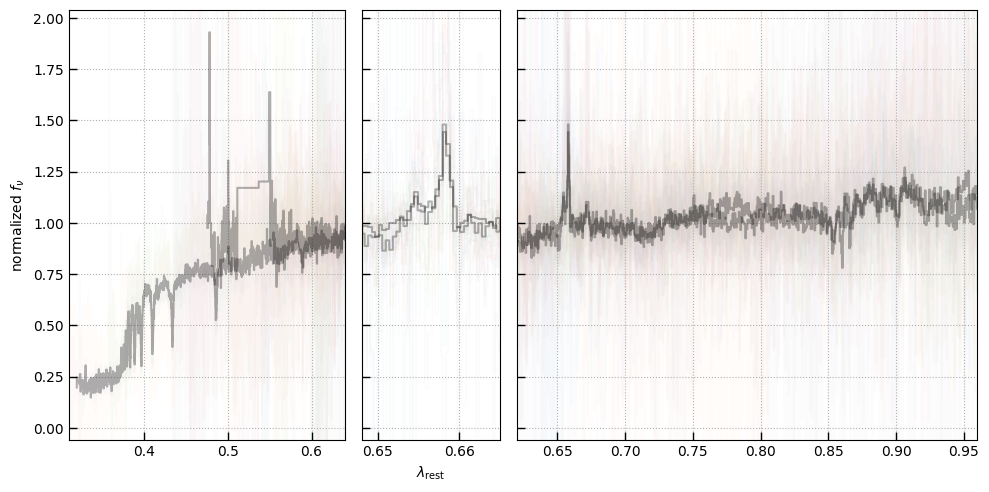
Grating line fluxes
Do some simple scatter plots of line ratios
Simple OIII 4959/5007
has_line = (
(tab['line_oiii_5007'] > 5 * tab['line_oiii_5007_err'])
& (tab['line_oiii_4959_err'] > 0)
& (tab['grade'] == 3)
& np.isin(tab['grating'], ['G140M', 'G235M', 'G395M'])
)
print(f'OIII in grating spectra: {has_line.sum()}')
ung = utils.Unique(tab['grating'], verbose=False)
fig, ax = plt.subplots(1,1,figsize=(10,5))
for grating in ung.values:
test = has_line & ung[grating]
if test.sum() == 0:
continue
ax.scatter(
tab['zrf'][test], (tab['line_oiii_4959'] / tab['line_oiii_5007'])[test],
alpha=0.1,
label=grating,
)
ax.hlines(1./2.98, *ax.get_xlim(), color='k', ls=":", label="OIII 5007/4959 = 2.98")
ax.legend(loc='upper left')
ax.set_ylim(0, 1)
ax.grid()
OIII in grating spectra: 7040
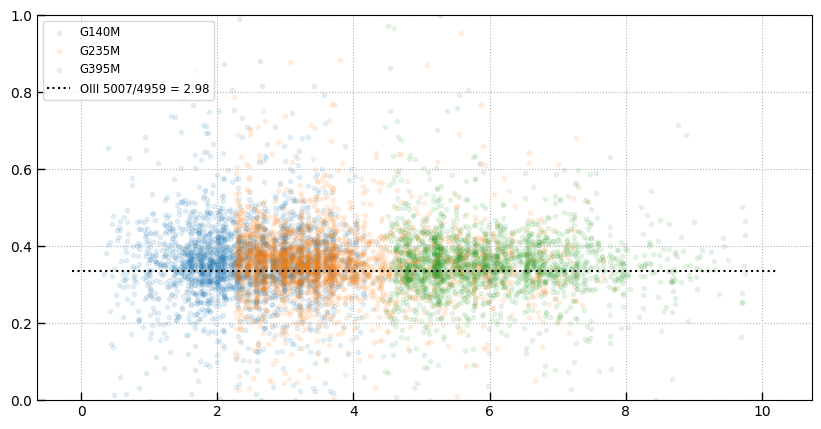
Stellar mass vs grating [NII] / H$\alpha$
Rough tracer of the mass-metallicity relation.
dz = np.abs(tab['zrf'] - tab['z_phot']) / (1+tab['zrf'])
has_line = (
(tab['line_ha'] > 3 * tab['line_ha_err'])
& (tab['line_nii_6584_err'] > 0)
& (tab['grade'] == 3)
& np.isin(tab['grating'], ['G140M', 'G235M', 'G395M'])
& (np.abs(dz) < 0.1)
)
print(f'Halpha in grating spectra: {has_line.sum()}')
fig, axes = plt.subplots(1,2,figsize=(8, 5), sharey=True)
scale_func = np.arcsinh
kws = dict(
c=np.log(1+tab['zrf'][has_line]),
cmap='rainbow',
vmin=np.log(1+1.0), vmax=np.log(1+7),
alpha=0.1
)
axes[0].scatter(
np.log10(tab['phot_mass'])[has_line],
scale_func(tab['line_nii_6584'] / tab['line_ha'])[has_line],
**kws
)
axes[1].scatter(
tab['zrf'][has_line],
scale_func(tab['line_nii_6584'] / tab['line_ha'])[has_line],
**kws
)
ax = axes[0]
yt = [-0.5, 0, 0.5] + list(range(6))[1:]
ax.set_yticks(scale_func(yt))
ax.set_yticklabels(yt)
ax.set_ylim(*scale_func([-0.6, 4]))
ax.set_xlim(5.9, 11.9)
ax.set_xlabel(r'$\log M/M_\odot$ (from photometry)')
ax.set_ylabel(r'[NII]$_{6584}$ / H$\alpha$')
axes[1].set_xlabel('redshift')
for ax in axes:
ax.grid()
fig.tight_layout(pad=1)
Halpha in grating spectra: 6476
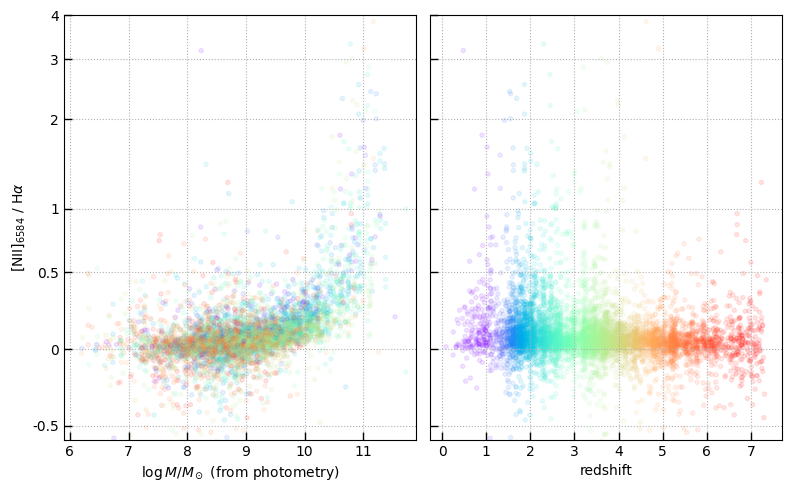
Thumbnail API
The DJA thumbnail API can create thumbnail figures and FITS cutouts of a requested set of filters at a particular coordinate.M
from IPython.display import Image
print(RGB_URL.format(**row))
Image(url=RGB_URL.format(**row), height=300, width=300)
https://grizli-cutout.herokuapp.com/thumb?size=1.5&scl=2.0&asinh=True&filters=f115w-clear%2Cf277w-clear%2Cf444w-clear&rgb_scl=1.5%2C0.74%2C1.3&pl=2&coord=214.91554591%2C52.94901831
print(SLIT_URL.format(**row))
Image(url=SLIT_URL.format(**row), height=300, width=300)
https://grizli-cutout.herokuapp.com/thumb?size=1.5&scl=4.0&invert=True&filters=f444w-clear&rgb_scl=1.5%2C0.74%2C1.3&pl=2&coord=214.91554591%2C52.94901831&nirspec=True&dpi_scale=6&nrs_lw=0.5&nrs_alpha=0.8&metafile=jw04233006001